Targeting Sodium Transport Reveals CHP1 Downregulation as a Novel Molecular Feature of Malignant Progression in Clear Cell Renal Cell Carcinoma: Insights from Integrated Multi-Omics Analyses
- PMID: 40723891
- PMCID: PMC12292469
- DOI: 10.3390/biom15071019
Targeting Sodium Transport Reveals CHP1 Downregulation as a Novel Molecular Feature of Malignant Progression in Clear Cell Renal Cell Carcinoma: Insights from Integrated Multi-Omics Analyses
Abstract
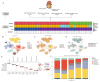
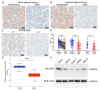
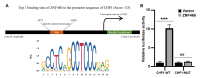
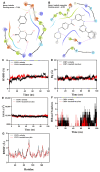
Clear cell renal cell carcinoma (ccRCC), the most common RCC subtype, displays significant intratumoral heterogeneity driven by metabolic reprogramming, which complicates our understanding of disease progression and limits treatment efficacy. This study aimed to construct a comprehensive cellular and transcriptional landscape of ccRCC, with emphasis on gene expression dynamics during malignant progression. An integrated analysis of 90 scRNA-seq samples comprising 534,227 cells revealed a progressive downregulation of sodium ion transport-related genes, particularly CHP1 (calcineurin B homologous protein isoform 1), which is predominantly expressed in epithelial cells. Reduced CHP1 expression was confirmed at both mRNA and protein levels using bulk RNA-seq, CPTAC proteomics, immunohistochemistry, and ccRCC cell lines. Survival analysis showed that high CHP1 expression correlated with improved prognosis. Functional analyses, including pseudotime trajectory, Mfuzz clustering, and cell-cell communication modeling, indicated that CHP1+ epithelial cells engage in immune interaction via PPIA-BSG signaling. Transcriptomic profiling and molecular docking suggested that CHP1 modulates amino acid transport through SLC38A1. ZNF460 was identified as a potential transcription factor of CHP1. Virtual screening identified arbutin and imatinib mesylate as candidate CHP1-targeting compounds. These findings establish CHP1 downregulation as a novel molecular feature of ccRCC progression and support its utility as a prognostic biomarker.
Keywords: calcineurin B homologous protein isoform 1; clear cell renal cell carcinoma; prognostic biomarker; single-cell RNA sequencing; sodium transport; tumor microenvironment.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Multi-omics analysis reveals the role of ribosome biogenesis in malignant clear cell renal cell carcinoma and the development of a machine learning-based prognostic model.Front Immunol. 2025 Jun 26;16:1602898. doi: 10.3389/fimmu.2025.1602898. eCollection 2025. Front Immunol. 2025. PMID: 40642093 Free PMC article.
-
Unraveling the Therapeutic Potential of Scutellarin for Clear Cell Renal Cell Carcinoma: A Comprehensive Molecular Analysis.Curr Pharm Des. 2025;31(25):2038-2062. doi: 10.2174/0113816128340451241224055536. Curr Pharm Des. 2025. PMID: 40265430
-
COL6A2 in clear cell renal cell carcinoma: a multifaceted driver of tumor progression, immune evasion, and drug sensitivity.J Transl Med. 2025 Aug 6;23(1):875. doi: 10.1186/s12967-025-06793-9. J Transl Med. 2025. PMID: 40770761 Free PMC article.
-
CYP1B1 promotes angiogenesis and sunitinib resistance in clear cell renal cell carcinoma via USP5-mediated HIF2α deubiquitination.Neoplasia. 2025 Aug;66:101186. doi: 10.1016/j.neo.2025.101186. Epub 2025 May 27. Neoplasia. 2025. PMID: 40435846 Free PMC article. Review.
-
A Systematic Review and Meta-analysis Comparing the Effectiveness and Adverse Effects of Different Systemic Treatments for Non-clear Cell Renal Cell Carcinoma.Eur Urol. 2017 Mar;71(3):426-436. doi: 10.1016/j.eururo.2016.11.020. Epub 2016 Dec 8. Eur Urol. 2017. PMID: 27939075
References
-
- Chen S., Cao Z., Prettner K., Kuhn M., Yang J., Jiao L., Wang Z., Li W., Geldsetzer P., Barnighausen T., et al. Estimates and Projections of the Global Economic Cost of 29 Cancers in 204 Countries and Territories From 2020 to 2050. JAMA Oncol. 2023;9:465–472. doi: 10.1001/jamaoncol.2022.7826. - DOI - PMC - PubMed
-
- Moch H., Amin M.B., Berney D.M., Comperat E.M., Gill A.J., Hartmann A., Menon S., Raspollini M.R., Rubin M.A., Srigley J.R., et al. The 2022 World Health Organization Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2022;82:458–468. doi: 10.1016/j.eururo.2022.06.016. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

