PHF20L1: An Epigenetic Regulator in Cancer and Beyond
- PMID: 40723918
- PMCID: PMC12292733
- DOI: 10.3390/biom15071048
PHF20L1: An Epigenetic Regulator in Cancer and Beyond
Abstract
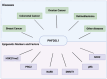
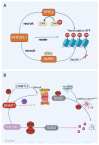
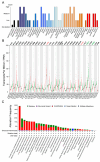
Plant homeodomain (PHD) finger protein 20-like 1 (PHF20L1) is a novel epigenetic "reader" that specifically recognises histone post-translational modifications (PTMs) via its Tudor and PHD finger domains, thereby regulating chromatin remodelling, DNA damage repair, and oncogene transcriptional activation. This review comprehensively summarises the role of PHF20L1 in various cancers, including breast, ovarian, and colorectal cancers, as well as retinoblastomas, and elucidates its molecular mechanisms of action in cancer pathogenesis. Accumulating evidence indicates that PHF20L1 is upregulated in these malignancies and drives tumour progression by promoting proliferation, metastasis, and immune evasion. Furthermore, PHF20L1 orchestrates tumour-related gene expression by interacting with key epigenetic complexes. Given its unique structural features, we propose novel strategies for developing small-molecule inhibitors and combinatorial therapies, providing a theoretical basis for targeted epigenetic regulation for precision treatment. Future research should further investigate the molecular regulatory networks of PHF20L1 in different cancers and other human diseases and focus on developing specific small-molecule inhibitors to enable precision-targeted therapies.
Keywords: PHF20L1; cancer; epigenetic regulation; histone methylation; targeted therapy.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
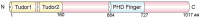
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

