Genetic Diversity in the Suppressyn Gene Sequence: From Polymorphisms to Loss-of-Function Mutations
- PMID: 40723922
- PMCID: PMC12293451
- DOI: 10.3390/biom15071051
Genetic Diversity in the Suppressyn Gene Sequence: From Polymorphisms to Loss-of-Function Mutations
Abstract
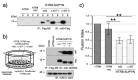
The suppressive regulator of cell fusion, suppressyn, is specifically expressed in the human placenta and is thought to play a crucial role in trophoblast fusion or syncytialization. Previous studies have suggested that alterations in its expression are associated with aberrant placental development, such as the immature placental morphology observed in Down syndrome, and may contribute to the pathogenesis of fetal growth restriction. While syncytialization in trophoblasts is an essential process for normal placental development, the precise molecular causes of its dysregulation remain poorly understood. In the present study, we aimed to elucidate the potential contribution of genomic variation to the loss of suppressyn function, extending previous analyses of expression abnormalities in perinatal disorders. Through sequence analysis, (1) we identified six polymorphisms within the coding region of the suppressyn gene, and (2) discovered that certain deletions and specific amino acid substitutions result in a complete loss of suppressyn-mediated inhibition of cell fusion. Although these mutations have not yet been reported in disease-associated genomic databases, our findings suggest that comprehensive genomic studies of perinatal and other disorders may reveal pathogenic variants of suppressyn, thereby uncovering novel genetic contributions to placental dysfunction. It is also anticipated that these findings might direct the development of therapeutic strategies targeting loss-of-function mutations.
Keywords: SNP; cell fusion; endogenous retroviruses (ERV); loss-of-function; suppressyn.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

