Mapping Integron-Associated AMR Genes in Whole Genome Sequences of Salmonella Typhimurium from Dairy Cattle
- PMID: 40723936
- PMCID: PMC12291841
- DOI: 10.3390/antibiotics14070633
Mapping Integron-Associated AMR Genes in Whole Genome Sequences of Salmonella Typhimurium from Dairy Cattle
Abstract
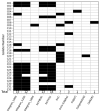
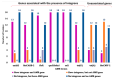
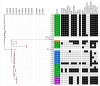
Background: Antimicrobial resistance (AMR) is a critical global health threat, with AMR Salmonella enterica serovar Typhimurium strains being a major foodborne pathogen. Integrons, a type of mobile genetic element, capture and transfer resistance genes, thereby playing a role in the spread of AMR. Objectives: This study aimed to characterize the locations of integrons carrying AMR genes within the whole genomes of 32 Salmonella Typhimurium isolates collected from dairy cattle by two U.S. Veterinary Diagnostic Laboratories between 2009 and 2012. Methods: Class I integrons were sequenced from PCR-amplified products. DNA was extracted, quantified, barcoded, and sequenced on the Illumina MiSeq platform. Whole genome sequences were trimmed and assembled using the SPAdes assembler in Geneious Prime®, and plasmids were identified with the PlasmidFinder pipeline in Linux. Integron locations were determined by aligning their sequences with whole genome contigs and plasmids, while AMR genes were identified through BLAST with the MEGARes 3.0 database and confirmed by alignment with isolate, plasmid, and integron sequences. Statistical analysis was applied to compare the proportions of isolates harboring integrons on their chromosome versus plasmids and also to examine the associations between integron presence and AMR gene presence. Results: Seven plasmid types were identified from all isolates: IncFII(S) (n = 14), IncFIB(S) (n = 13), IncC (n = 7), Inc1-I(Alpha) (n = 3), and ColpVC, Col(pAHAD28), and Col8282 (1 isolate each). Of the 32 isolates, 16 (50%) carried at least one size of integron. Twelve of them carried both 1000 and 1200 bp; 3 carried only 1000 bp and 1 carried 1800 bp integrons. Of the 15 isolates that carried 1000 bp integron, 12 harbored it on IncFIB(S) plasmids, 2 on IncC plasmids, and 1 on the chromosome. The 1200 bp integrons from all 12 isolates were located on chromosomes. There were significant positive associations between the presence of integrons and the presence of several AMR genes including sul1, aadA2, blaCARB-2, qacEdelta1, tet(G), and floR (p < 0.05). AMR genes were located as follows: aadA2 on IncFIB(S) and IncC plasmids; blaCMY-2 on IncC plasmid; qacEdelta1 on IncFIB(S), IncC, and chromosome; blaCARB-2, floR, tet(A) and tet(G) on the chromosome. Conclusions: The findings highlight the genomic and plasmid complexity of Salmonella Typhimurium which is impacted by the presence and location of integrons, and this study provides genomic insights that can inform efforts to enhance food safety and protect both animal and public health.
Keywords: Salmonella Typhimurium; antimicrobial resistance genes; cattle; class-1 integrons; genomic location; plasmids; whole genome sequencing.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

References
-
- World Health Organization . WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance, to Guide Research, Development, and Strategies to Prevent and Control Antimicrobial Resistance. World Health Organization; Geneva, Switzerland: 2024.
-
- Ahmed S.K., Hussein S., Qurbani K., Ibrahim R.H., Fareeq A., Mahmood K.A., Mohamed M.G. Antimicrobial Resistance: Impacts, Challenges, and Future Prospects. J. Med. Surg. Public Health. 2024;2:100081. doi: 10.1016/j.glmedi.2024.100081. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

