First Report of Stenotrophomonas maltophilia from Canine Dermatological Infections: Unravelling Its Antimicrobial Resistance, Biofilm Formation, and Virulence Traits
- PMID: 40723942
- PMCID: PMC12291639
- DOI: 10.3390/antibiotics14070639
First Report of Stenotrophomonas maltophilia from Canine Dermatological Infections: Unravelling Its Antimicrobial Resistance, Biofilm Formation, and Virulence Traits
Abstract
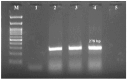
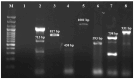
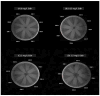
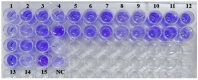
Background/Objectives: The present study was aimed at documenting S. maltophilia occurrence in dogs with skin ailments, investigating its virulence, biofilm-forming ability, antimicrobial susceptibility, and zoonotic potential to inform preventive and therapeutic strategies against multidrug resistant S. maltophilia infections. Methods: Skin swabs (n = 300) were collected from dogs with dermatological ailments. Isolation was performed using selective media and confirmed with molecular methods, validated by MALDI Biotyper. Antimicrobial susceptibility testing and efflux activity assessment were conducted. Resistance genes related to sulfonamides, quinolones, and β-lactams were screened. Virulence was assessed by biofilm formation, motility, and virulence gene profiling. Results: In total, 15 S. maltophilia (5%) isolates were identified. All 15 isolates were susceptible to trimethoprim-sulfamethoxazole, enrofloxacin, gatifloxacin, levofloxacin, minocycline, and tigecycline, but resistant to cefpodoxime and aztreonam. The following resistance genes qnr (93.3%), blaOXA-48 (46.7%), blaKPC (33.3%), blaNDM (33.3%), blaCTX-M (20%), blaSHV (20%), and blaTEM (6.7%) were detected. All 15 isolates displayed high efflux activity. Overall, 9 isolates (60%) were strong biofilm producers, and 6 (40%) were moderate. Virulence genes such as virB, motA, rmlA, and fliC were present in all 15 isolates, with others varying in frequency. All isolates exhibited swimming motility. Heat map clustering showed diverse profiles, with no identical isolate patterns. Correlation analysis indicated positive associations between several antimicrobial resistance and virulence genes. Conclusions: This study underscores the zoonotic potential of S. maltophilia from dogs, advocating for a One Health approach to mitigate infection risks and limit the spread of virulent multidrug resistant pathogens.
Keywords: One Health; dogs; skin swabs; stenotrophomonas maltophilia; zoonotic.
Conflict of interest statement
Author Charley A. Cull was employed by Midwest Veterinary Services, Inc., Oakland, Nebraska, 68045, USA. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest. The funders had no role in the design of this study; in the collection, analyses, or interpretation of data; in the writing of this manuscript; or in the decision to publish the results.
Figures



Similar articles
-
Pan-drug, colistin, streptomycin, erythromycin, clindamycin resistant Salmonella enterica serovars isolated from slaughtered cattle and human in mansoura, Egypt.Ann Clin Microbiol Antimicrob. 2025 Jul 3;24(1):40. doi: 10.1186/s12941-025-00809-4. Ann Clin Microbiol Antimicrob. 2025. PMID: 40611300 Free PMC article.
-
Antibiotic resistance, biofilm formation, and virulence gene profiling of multidrug-resistant Pseudomonas aeruginosa isolates from a multi-speciality hospital in Sikkim, India.Microb Pathog. 2025 Aug 7;208:107976. doi: 10.1016/j.micpath.2025.107976. Online ahead of print. Microb Pathog. 2025. PMID: 40782980
-
Retrospective analysis of antibiotic resistance profiles and frequency of resistance genes in clinical Stenotrophomonas maltophilia isolates.Acta Microbiol Immunol Hung. 2025 May 22;72(2):139-144. doi: 10.1556/030.2025.02582. Print 2025 Jun 20. Acta Microbiol Immunol Hung. 2025. PMID: 40402576
-
Global prevalence and distribution of antibiotic resistance among clinical isolates of Stenotrophomonas maltophilia: A systematic review and meta-analysis.J Glob Antimicrob Resist. 2023 Sep;34:253-267. doi: 10.1016/j.jgar.2023.02.018. Epub 2023 Mar 9. J Glob Antimicrob Resist. 2023. PMID: 36906172
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
LinkOut - more resources
Full Text Sources

