New 1,2,4-Triazole Derivatives with a N-Mannich Base Structure Based on a 4,6-Dimethylpyridine Scaffold as Anticancer Agents: Design, Synthesis, Biological Evaluation, and Molecular Modeling
- PMID: 40724822
- PMCID: PMC12294558
- DOI: 10.3390/ijms26146572
New 1,2,4-Triazole Derivatives with a N-Mannich Base Structure Based on a 4,6-Dimethylpyridine Scaffold as Anticancer Agents: Design, Synthesis, Biological Evaluation, and Molecular Modeling
Abstract
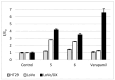
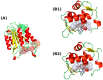
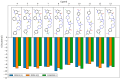
A series of novel N-Mannich bases derived from a dimethylpyridine-1,2,4-triazole hybrid was synthesized and evaluated in vitro for cytotoxic activity on several human gastrointestinal cancer cells (EPG, Caco-2, LoVo, LoVo/Dx, and HT-29). Compound 6 bearing a phenyl group at the N-4 position and a 4-methylphenyl piperazine moiety at the N-2 position of the 1,2,4-triazole-3-thione scaffold exerted good cytotoxic activities on EPG and Caco-2 cell lines, along with pronounced selectivity, showing lower cytotoxicity against normal colonic epithelial cells (CCD 841 CoTr). Further evaluation revealed the good ability of compound 6 to inhibit the efflux function of P-glycoprotein in P-gp-expressing cell lines (HT-29, LoVo, and LoVo/Dx). Moreover, compound 6 induced apoptotic cell death through a significant increase in the caspase-3 and p53 protein levels in HT-29 cells. Finally, the molecular docking method was applied to explain our experimental findings. The molecular modeling study based on Density Functional Theory (DFT) and the Quantum Theory of Atoms in Molecules (QTAIM) analysis provided insight into the geometric and electronic structure properties of the compounds.
Keywords: 1,2,4-triazole; DFT; N-Mannich base; QTAIM; anticancer activity; cytotoxicity; dimethylpyridine; molecular docking.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








Similar articles
-
Novel purine-linked 1,2,3-triazole derivatives as effective anticancer agents: design, synthesis, docking, DFT, and ADME-T investigations.Sci Rep. 2025 Jul 23;15(1):26853. doi: 10.1038/s41598-025-95669-5. Sci Rep. 2025. PMID: 40702234 Free PMC article.
-
Efficient Synthesis, Anticancer Evaluation of Triazole-Thiadiazole/Benzo[d]Oxazole Scaffolds, and Investigation of Their Reactivity Properties Using Density-Functional Theory Calculations and In Silico Docking.Chem Biodivers. 2025 Jul;22(7):e202402470. doi: 10.1002/cbdv.202402470. Epub 2025 Mar 11. Chem Biodivers. 2025. PMID: 39972950
-
New Schiff bases derived from dimethylpyridine-1,2,4-triazole hybrid as cytotoxic agents targeting gastrointestinal cancers: Design, synthesis, biological evaluation and molecular docking studies.Bioorg Chem. 2023 Oct;139:106758. doi: 10.1016/j.bioorg.2023.106758. Epub 2023 Jul 31. Bioorg Chem. 2023. PMID: 37540951
-
Recent Advances in 1,2,4-Triazole-Based Anticancer Agents: Structural Optimization, Mechanisms, and Therapeutic Potential (2022-2025).Arch Pharm (Weinheim). 2025 Jul;358(7):e70059. doi: 10.1002/ardp.70059. Arch Pharm (Weinheim). 2025. PMID: 40726245 Review.
-
Therapeutic potential of chalcone-1,2,3-triazole hybrids as anti-tumour agents: a systematic review and SAR studies.Future Med Chem. 2025 Feb;17(4):449-465. doi: 10.1080/17568919.2025.2458450. Epub 2025 Jan 31. Future Med Chem. 2025. PMID: 39886772
References
-
- Shabelnyk K., Fominichenko A., Antypenko O., Gaponov O., Koptieva S., Shyshkina S., Voskoboinik O., Okovytyy S., Kovalenko S., Oksenych V., et al. Antistaphylococcal Triazole-Based Molecular Hybrids: Design, Synthesis and Activity. Pharmaceuticals. 2025;18:83. doi: 10.3390/ph18010083. - DOI - PMC - PubMed
-
- Zaheer M., Zia-Ur-Rehman M., Munir R., Jamil N., Ishtiaq S., Zaib Saleem R.S., Elsegood M.R.J. (Benzylideneamino)Triazole-Thione Derivatives of Flurbiprofen: An Efficient Microwave-Assisted Synthesis and in Vivo Analgesic Potential. ACS Omega. 2021;6:31348–31357. doi: 10.1021/acsomega.1c05222. - DOI - PMC - PubMed
-
- Nawaz Z., Riaz N., Saleem M., Iqbal A., Abida Ejaz S., Bashir B., Muzaffar S., Ashraf M., Aziz-ur-Rehman, Sajjad Bilal M., et al. Molecular Hybrids of Substituted Phenylcarbamoylpiperidine and 1,2,4-Triazole Methylacetamide as Potent 15-LOX Inhibitors: Design, Synthesis, DFT Calculations and Molecular Docking Studies. Bioorg. Chem. 2024;143:106984. doi: 10.1016/j.bioorg.2023.106984. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

