Comparing DNA Methylation Landscapes in Peripheral Blood from Myalgic Encephalomyelitis/Chronic Fatigue Syndrome and Long COVID Patients
- PMID: 40724879
- PMCID: PMC12294161
- DOI: 10.3390/ijms26146631
Comparing DNA Methylation Landscapes in Peripheral Blood from Myalgic Encephalomyelitis/Chronic Fatigue Syndrome and Long COVID Patients
Abstract
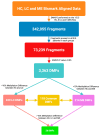
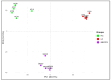
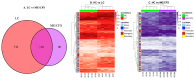
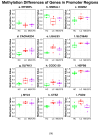
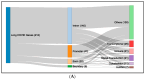
Post-viral conditions, Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) and Long COVID (LC), share > 95% of their symptoms, but the connection between disturbances in their underlying molecular biology is unclear. This study investigates DNA methylation patterns in peripheral blood mononuclear cells (PBMC) from patients with ME/CFS, LC, and healthy controls (HC). Reduced Representation Bisulphite Sequencing (RRBS) was applied to the DNA of age- and sex-matched cohorts: ME/CFS (n = 5), LC (n = 5), and HC (n = 5). The global DNA methylomes of the three cohorts were similar and spread equally across all chromosomes, except the sex chromosomes, but there were distinct minor changes in the exons of the disease cohorts towards more hypermethylation. A principal component analysis (PCA) analysing significant methylation changes (p < 0.05) separated the ME/CFS, LC, and HC cohorts into three distinct clusters. Analysis with a limit of >10% methylation difference and at p < 0.05 identified 214 Differentially Methylated Fragments (DMF) in ME/CFS, and 429 in LC compared to HC. Of these, 118 DMFs were common to both cohorts. Those in promoters and exons were mainly hypermethylated, with a minority hypomethylated. There were rarer examples with either no change in methylation in ME/CFS but a change in LC, or a methylation change in ME/CFS but in the opposite direction in LC. The differential methylation in a number of fragments was significantly greater in the LC cohort than in the ME/CFS cohort. Our data reveal a generally shared epigenetic makeup between ME/CFS and LC but with specific, distinct changes. Differences between the two cohorts likely reflect the stage of the disease from onset (LC 1 year vs. ME/CFS 12 years), but specific changes imposed by the SARS-CoV-2 virus in the case of the LC patients cannot be discounted. These findings provide a foundation for further studies with larger cohorts at the same disease stage and for functional analyses to establish clinical relevance.
Keywords: DNA methylation; Myalgic Encephalomyelitis/Chronic Fatigue Syndrome; differentially methylated fragments; hypermethylation; hypomethylation; long COVID; peripheral blood mononuclear cells.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Elevated risk of new-onset chronic fatigue syndrome/myalgic encephalomyelitis up to four years after SARS-CoV-2 infection.J Transl Med. 2025 Jul 23;23(1):815. doi: 10.1186/s12967-025-06625-w. J Transl Med. 2025. PMID: 40702518 Free PMC article.
-
Recovery from Exercise in Persons with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS).Medicina (Kaunas). 2023 Mar 15;59(3):571. doi: 10.3390/medicina59030571. Medicina (Kaunas). 2023. PMID: 36984572 Free PMC article.
-
Changes in DNA methylation profiles of myalgic encephalomyelitis/chronic fatigue syndrome patients reflect systemic dysfunctions.Clin Epigenetics. 2020 Nov 4;12(1):167. doi: 10.1186/s13148-020-00960-z. Clin Epigenetics. 2020. PMID: 33148325 Free PMC article.
-
Health-related quality of life in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome and Post COVID-19 Condition: a systematic review.J Transl Med. 2025 Mar 13;23(1):318. doi: 10.1186/s12967-025-06131-z. J Transl Med. 2025. PMID: 40075382 Free PMC article.
-
Exercise therapy for chronic fatigue syndrome.Cochrane Database Syst Rev. 2024 Dec 19;12(12):CD003200. doi: 10.1002/14651858.CD003200.pub9. Cochrane Database Syst Rev. 2024. PMID: 39697147
References
-
- WHO COVID-19 Cases, World. 2025. [(accessed on 3 April 2025)]. Available online: https://data.who.int/dashboards/covid19/cases.
-
- National Academies of Sciences, Engineering, and Medicine. Health and Medicine Division. Board on Global Health. Board on Health Sciences Policy. Committee on Examining the Working Definition for Long COVID . In: A Long COVID Definition: A Chronic, Systemic Disease State with Profound Consequences. Goldowitz I., Worku T., Brown L., Fineberg H.V., editors. National Academies Press (US); Washington, DC, USA: 2024. - PubMed
-
- Collins F.S. NIH Launches New Initiative to Study “Long COVID”. [(accessed on 1 April 2025)];2021 Available online: https://www.nih.gov/about-nih/who-we-are/nih-director/statements/nih-lau....
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

