Identification of Genes Linked to Meniscal Degeneration in Osteoarthritis: An In Silico Analysis
- PMID: 40724902
- PMCID: PMC12294481
- DOI: 10.3390/ijms26146651
Identification of Genes Linked to Meniscal Degeneration in Osteoarthritis: An In Silico Analysis
Abstract
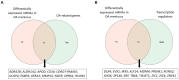
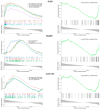
Meniscal degradation is considered a driver of osteoarthritis (OA) progression, but the underlying mechanisms leading to age-related meniscus degeneration remain unknown. This study aimed to identify key genes and pathways involved in meniscal degradation through a computational analysis. Gene expression profiles were obtained from the Gene Expression Omnibus (GEO) database. Differential expression gene (DEG) analysis was performed using DESeq2 accompanied by functional enrichment analysis, protein-protein interaction (PPI) and clustering analysis. Additionally, gene set enrichment analysis (GSEA) was performed. A total of 85 mRNAs (DEMs) and 8 long non-coding RNAs (DE LncRNAs) were found to be differentially expressed in OA meniscus tissues. Among 85 DEMs, 12 genes were found to be known OA-related genes, whereas 15 genes acted as transcription regulators, including RUNX2 and TBX4, which were identified as effector genes for OA. Enrichment analysis revealed the implication of DEMs in cartilage-degradation-related processes, including inflammatory pathways, lipid metabolism, extracellular matrix organization and superoxide/nitric oxide metabolic processes. Target genes of DE lncRNAs were found to be involved in chondrocyte differentiation and pathways related to cartilage degradation. A comparative analysis of meniscus, synovium and cartilage datasets identified three genes (GJB2, PAQR5 and CLEC12A) as being differentially expressed across all three OA-affected tissues, which were implicated in inflammatory and cholesterol metabolism processes. Our results support that shared mechanisms lead to meniscal and cartilage degradation during OA progression, providing further insights into the processes underlying OA pathogenesis and potential therapeutic targets for knee OA.
Keywords: in silico analysis; meniscus; osteoarthritis; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

