Repeatome Analysis of Plasma Circulating DNA in Patients with Cardiovascular Disease: Variation with Cell-Free DNA Integrity/Length and Clinical Parameters
- PMID: 40724907
- PMCID: PMC12294208
- DOI: 10.3390/ijms26146657
Repeatome Analysis of Plasma Circulating DNA in Patients with Cardiovascular Disease: Variation with Cell-Free DNA Integrity/Length and Clinical Parameters
Abstract
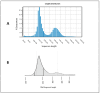
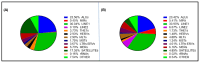
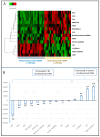
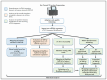
Repetitive DNA represents over 50% of the human genome and is an abundant component of circulating cell-free DNA (cfDNA). We previously showed that cfDNA levels and integrity can predict survival in elderly patients with cardiovascular disease. Here, we aimed to clarify whether a low-pass next-generation sequencing (NGS) approach can characterize the repeat content of cfDNA. Considering the bimodal distribution of cfDNA fragment lengths, we examined the occurrence of repetitive DNA subfamilies separately in dinucleosomal (>250 bp) and mononucleosomal (≤250 bp) cfDNA sequences from 24 patients admitted for heart failure. An increase in the relative abundance of Alu repetitive elements was observed in the longer fraction, while alpha satellites were enriched in the mononucleosomal fraction. The relative abundance of Alu, ALR, and L1HS DNA in the dinucleosomal fraction correlated with different prognostic biomarkers, and Alu DNA was negatively associated with the presence of chronic kidney disease comorbidity. These results, together with the observed inverse correlation between Alu DNA abundance and cfDNA integrity, suggest that the composition of plasma cfDNA could be determined by multiple mechanisms in different physio-pathological conditions. In conclusion, low-pass NGS is an inexpensive method to analyze the cfDNA repeat landscape and identify new cardiovascular disease biomarkers.
Keywords: Alu elements; CKD; cardiovascular disease; cfDNA; circulating biomarkers; eGFR; hearth failure; low-pass NGS; repetitive DNA.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Standardized Workflow and Analytical Validation of Cell-Free DNA Extraction for Liquid Biopsy Using a Magnetic Bead-Based Cartridge System.Cells. 2025 Jul 11;14(14):1062. doi: 10.3390/cells14141062. Cells. 2025. PMID: 40710315 Free PMC article.
-
Characterization of methylation profile in biofluid cell-free DNA and identification of differentially methylated genes for phenotypic representations in Parkinson's disease.Clin Neurol Neurosurg. 2025 Sep;256:109014. doi: 10.1016/j.clineuro.2025.109014. Epub 2025 Jun 10. Clin Neurol Neurosurg. 2025. PMID: 40527220
-
Cell-Free DNA in Metastatic Colorectal Cancer: A Systematic Review and Meta-Analysis.Oncologist. 2017 Sep;22(9):1049-1055. doi: 10.1634/theoncologist.2016-0178. Epub 2017 Aug 4. Oncologist. 2017. PMID: 28778958 Free PMC article.
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

