Comparative Transcriptome and Hormonal Analysis Reveals the Mechanisms of Salt Tolerance in Rice
- PMID: 40724910
- PMCID: PMC12295843
- DOI: 10.3390/ijms26146660
Comparative Transcriptome and Hormonal Analysis Reveals the Mechanisms of Salt Tolerance in Rice
Abstract
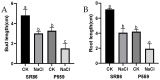
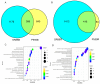
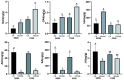
Salt stress is a major constraint to seed germination and early seedling growth in rice, affecting crop establishment and productivity. To understand the mechanisms underlying salt tolerance, we investigated two rice varieties with contrasting responses as follows: salt-tolerant sea rice 86 (SR86) and salt-sensitive P559. Germination assays under increasing NaCl concentrations (50-300 mM) revealed that 100 mM NaCl induced clear phenotypic divergence. SR86 maintained bud growth and showed enhanced root elongation under moderate salinity, while P559 exhibited significant growth inhibition. Transcriptomic profiling of buds and roots under 100 mM NaCl identified over 3724 differentially expressed genes (DEGs), with SR86 showing greater transcriptional plasticity, particularly in roots. Gene ontology enrichment revealed tissue- and genotype-specific responses. Buds showed enrichment in photosynthesis-related and redox-regulating pathways, while roots emphasized ion transport, hormonal signaling, and oxidative stress regulation. SR86 specifically activated genes related to photosystem function, DNA repair, and transmembrane ion transport, while P559 showed activation of oxidative stress-related and abscisic acid (ABA)-regulated pathways. Hormonal profiling supported transcriptomic findings as follows: both varieties showed increased gibberellin 3 (GA3) and gibberellin 4 (GA4) levels under salt stress. SR86 showed elevated auxin (IAA) and reduced jasmonic acid (JA), whereas P559 maintained stable IAA and JA levels. Ethylene precursor and salicylic acid levels declined in both varieties. ABA levels rose slightly but not significantly. These findings suggest that SR86's superior salt tolerance results from rapid growth, robust transcriptional reprogramming, and coordinated hormonal responses. This study offers key insights into early-stage salt stress adaptation and identifies molecular targets for improving stress resilience in rice.
Keywords: hormonal regulation; rice; salt stress; salt-tolerance mechanisms; seed germination; transcriptome analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
OsWNK9 regulates the expression of key transcription factors, phytohormonal, and transporters genes to improve salinity stress tolerance in rice.Sci Rep. 2025 Aug 22;15(1):30930. doi: 10.1038/s41598-025-14775-6. Sci Rep. 2025. PMID: 40847042
-
Combining QTL mapping and transcriptomics to identify candidate genes for cold tolerance during the budding and seedling stages in rice.BMC Genomics. 2025 Aug 19;26(1):756. doi: 10.1186/s12864-025-11937-8. BMC Genomics. 2025. PMID: 40830828 Free PMC article.
-
Screening of salt-tolerant soybean germplasm and study of salt-tolerance mechanism.Plant Cell Rep. 2025 Aug 1;44(8):187. doi: 10.1007/s00299-025-03574-y. Plant Cell Rep. 2025. PMID: 40748503
-
Salinity Stress in Rice: Multilayered Approaches for Sustainable Tolerance.Int J Mol Sci. 2025 Jun 23;26(13):6025. doi: 10.3390/ijms26136025. Int J Mol Sci. 2025. PMID: 40649804 Free PMC article. Review.
-
Promotion of Ca2+ Accumulation in Roots by Exogenous Brassinosteroids as a Key Mechanism for Their Enhancement of Plant Salt Tolerance: A Meta-Analysis and Systematic Review.Int J Mol Sci. 2023 Nov 9;24(22):16123. doi: 10.3390/ijms242216123. Int J Mol Sci. 2023. PMID: 38003311 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- KJRC2023D13 and KJRC2023B23/Innovational Fund for Scientific and Technological Personnel of Hainan Province
- SCKJ-JYRC-2023-30/Project of Sanya Yazhou Bay Science and Technology City
- ZDKJ202001/Hainan Province Major Science and Technology project Fund
- HAAS2023RCQD16/Project of Hainan Academy of Agricultural Sciences
LinkOut - more resources
Full Text Sources

