FR-BINN: Biologically Informed Neural Networks for Enhanced Biomarker Discovery and Pathway Analysis
- PMID: 40724918
- PMCID: PMC12294759
- DOI: 10.3390/ijms26146670
FR-BINN: Biologically Informed Neural Networks for Enhanced Biomarker Discovery and Pathway Analysis
Abstract
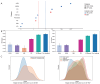
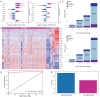
Chronic inflammation plays a pivotal role in human health, with certain inflammatory conditions significantly increasing the risk of cancer, while others do not. However, the molecular mechanisms underlying this divergent risk remain poorly understood. In this study, we propose FR-BINN, a biologically informed neural network framework for disease prediction and interpretability. Incorporating Fenton reaction (FR)-related biological priors and leveraging multiple interpretability methods, FR-BINN identifies key genes driving cancer-prone and non-cancer-prone chronic inflammatory diseases. The experimental results demonstrate that FR-BINN achieves superior classification performance while offering biologically interpretable insights. Moreover, attribution results derived from different explainable techniques show high consistency, and intra-method results exhibit distinct patterns across disease categories. We further combine large language models with feature attributions to identify candidate biomarkers, and independent datasets confirm the robustness of these findings. Notably, genes such as NCOA1 and SDHB are identified as being associated with cancer susceptibility. The framework further reveals distinct patterns in energy metabolism, oxidative stress, and pH regulation between cancer-prone and non-cancer-prone inflammatory diseases. These insights enhance our understanding of inflammation-associated tumorigenesis and contribute to the identification of potential biomarkers and therapeutic targets.
Keywords: biologically informed neural network; biomarker; chronic inflammation; explainable artificial intelligence.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
A Responsible Framework for Assessing, Selecting, and Explaining Machine Learning Models in Cardiovascular Disease Outcomes Among People With Type 2 Diabetes: Methodology and Validation Study.JMIR Med Inform. 2025 Jun 27;13:e66200. doi: 10.2196/66200. JMIR Med Inform. 2025. PMID: 40577645 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
How lived experiences of illness trajectories, burdens of treatment, and social inequalities shape service user and caregiver participation in health and social care: a theory-informed qualitative evidence synthesis.Health Soc Care Deliv Res. 2025 Jun;13(24):1-120. doi: 10.3310/HGTQ8159. Health Soc Care Deliv Res. 2025. PMID: 40548558
-
Stabilizing machine learning for reproducible and explainable results: A novel validation approach to subject-specific insights.Comput Methods Programs Biomed. 2025 Sep;269:108899. doi: 10.1016/j.cmpb.2025.108899. Epub 2025 Jun 21. Comput Methods Programs Biomed. 2025. PMID: 40570739
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Rocca W.A., Petersen R.C., Knopman D.S., Hebert L.E., Evans D.A., Hall K.S., Gao S., Unverzagt F.W., Langa K.M., Larson E.B., et al. Trends in the incidence and prevalence of Alzheimer’s disease, dementia, and cognitive impairment in the United States. Alzheimer’s Dement. 2011;7:80–93. doi: 10.1016/j.jalz.2010.11.002. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

