In Silico Analysis of Post-COVID-19 Condition (PCC) Associated SNP rs9367106 Predicts the Molecular Basis of Abnormalities in the Lungs and Brain Functions
- PMID: 40724930
- PMCID: PMC12294317
- DOI: 10.3390/ijms26146680
In Silico Analysis of Post-COVID-19 Condition (PCC) Associated SNP rs9367106 Predicts the Molecular Basis of Abnormalities in the Lungs and Brain Functions
Abstract
Long- or post-COVID-19 syndrome, which is also designated by WHO as Post COVID-19 Condition (PCC), is characterized by the persistent symptoms that remain after recovery from SARS-CoV-2 infection. A worldwide consortium of Long COVID-19 Host Genetics Initiative (Long COVID-19 HGI) identified an SNP rs9367106 (G>C; chr6:41,515,652, GRCh38, p = 1.76 × 10-10, OR = 1.63, 95% CI: 1.40-1.89) that is associated with PCC. Unraveling the functional significance of this SNP is of prime importance to understanding the development of the PCC phenotypes and their therapy. Here, in Silico, I explored how the risk allele of this SNP alters the functional mechanisms and molecular pathways leading to the development of PCC phenotypes. Bioinformatic methods include physical interactions using HI-C and Chia-PET analysis, Transcription Factors (TFs) binding ability, RNA structure modeling, epigenetic, and pathway analysis. This SNP resides within two long RNA genes, LINC01276 and FOXP4-AS1, and is located at ~31 kb upstream of a transcription factor FOXP4. This DNA region, including this SNP, physically interacts with FOXP4-AS1 and FOXP4, implying that this regulatory SNP could alter the normal cellular function of FOXP4-AS1 and FOXP4. Furthermore, rs9367106 is in eQTL with the FOXP4 gene in lung tissue. rs9367106 carrying DNA sequences act as distant enhancers and bind with several transcription factors (TFs) including YY1, PPAR-α, IK-1, GR-α, and AP2αA. The G>C transition extensively modifies the RNA structure that may affect the TF bindings and enhancer functions to alter the interactions and functions of these RNA molecules. This SNP also includes an ALU/SINE sequence and alteration of which by the G>C transition may prevent IFIH1/MDA5 activation, leading to suppression of host innate immune responses. LINC01276 targets the MED20 gene that expresses mostly in brain tissues, associated with sleep disorders and basal ganglia abnormalities similar to some of the symptoms of PCC phenotypes. Taken together, G>C transition of rs9367601 may likely alter the function of all three genes to explain the molecular basis of developing the long-term symptomatic abnormalities in the lungs and brain observed after COVID-19 recovery.
Keywords: FOXP4; LINC01276; enhancer; polymorphism; post-COVID-19 condition; sleep disorder; transcription factor.
Conflict of interest statement
The author declares no conflicts of interest.
Figures
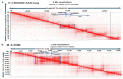
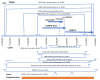

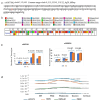
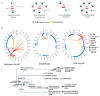
 and interacts with FOXP4
and interacts with FOXP4
 . The purple color
. The purple color  is the antisense FOXP4-AS1. (c) Mechanism of altered regulatory function by G>C transition of the SNP rs9367106. LINC01276 and FOXP4-AS1 RNA interact with FOXP4, and LINC01276 also interact with MED20 to develop PCC phenotypes in neuronal and lung tissues.
is the antisense FOXP4-AS1. (c) Mechanism of altered regulatory function by G>C transition of the SNP rs9367106. LINC01276 and FOXP4-AS1 RNA interact with FOXP4, and LINC01276 also interact with MED20 to develop PCC phenotypes in neuronal and lung tissues.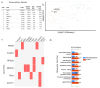
Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Multidisciplinary collaborative guidance on the assessment and treatment of patients with Long COVID: A compendium statement.PM R. 2025 Jun;17(6):684-708. doi: 10.1002/pmrj.13397. Epub 2025 Apr 22. PM R. 2025. PMID: 40261198
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
References
-
- Appelman B., Charlton B.T., Goulding R.P., Kerkhoff T.J., Breedveld E.A., Noort W., Offringa C., Bloemers F.W., van Weeghel M., Schomakers B.V., et al. Muscle abnormalities worsen after post-exertional malaise in long COVID. Nat. Commun. 2024;15:17. doi: 10.1038/s41467-023-44432-3. - DOI - PMC - PubMed
-
- Cervia-Hasler C., Brüningk S.C., Hoch T., Fan B., Muzio G., Thompson R.C., Ceglarek L., Meledin R., Westermann P., Emmenegger M., et al. Persistent complement dysregulation with signs of thromboinflammation in active Long Covid. Science. 2024;383:eadg7942. doi: 10.1126/science.adg7942. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

