Lipidome Complexity in Physiological and Pathological Skin Pigmentation
- PMID: 40725029
- PMCID: PMC12295737
- DOI: 10.3390/ijms26146785
Lipidome Complexity in Physiological and Pathological Skin Pigmentation
Abstract
Skin pigmentation results from complex cellular interactions and is influenced by genetic, environmental, and metabolic factors. Emerging evidence highlights the multiple pathways by which lipids regulate melanogenesis and points to lipid metabolism and signaling as key players in this process. Lipidomics is a high-throughput omics approach that enables detailed characterization of lipid profiles, thus representing a valid tool for evaluating skin lipid functional role in both physiological melanogenesis and pigmentary disorders. The use of lipidomics to gain a deeper comprehension of the role of lipids in skin pigmentation is still an evolving field, but it has allowed the identification of significant lipid dysregulation in several pigmentary pathologies. This review summarizes the current knowledge on the involvement of lipids in skin pigmentation, focusing on lipid profile alterations described in hyper- and hypopigmentary disorders such as post-inflammatory hyperpigmentation, melasma, solar lentigo, and vitiligo. Lipidomic profiling reveals disease-specific alterations supporting the pivotal role of lipid signaling in the physiopathological mechanisms of melanogenesis. These findings provide insights into disease pathogenesis and show promise for the discovery of biomarkers and innovative therapeutic strategies for pigmentary disorders.
Keywords: lipidomics; lipids; melanin; melanogenesis; melasma; skin; solar lentigo; vitiligo.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
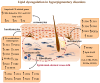
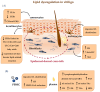
References
-
- Gutierrez Reyes C.D., Alejo-Jacuinde G., Perez Sanchez B., Chavez Reyes J., Onigbinde S., Mogut D., Hernández-Jasso I., Calderón-Vallejo D., Quintanar J.L., Mechref Y. Multi Omics Applications in Biological Systems. Curr. Issues Mol. Biol. 2024;46:5777–5793. doi: 10.3390/cimb46060345. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

