Detection of Feline Coronavirus Membrane Gene Based on Conventional Revere Transcription-Polymerase Chain Reaction, Nested Reverse Transcription-Polymerase Chain Reaction, and Reverse Transcription-Quantitative Polymerase Chain Reaction: A Comparative Study
- PMID: 40725108
- PMCID: PMC12295316
- DOI: 10.3390/ijms26146861
Detection of Feline Coronavirus Membrane Gene Based on Conventional Revere Transcription-Polymerase Chain Reaction, Nested Reverse Transcription-Polymerase Chain Reaction, and Reverse Transcription-Quantitative Polymerase Chain Reaction: A Comparative Study
Abstract
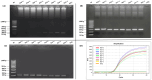
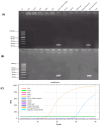
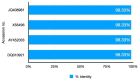
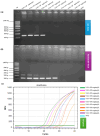
Feline coronavirus (FCoV) is a major pathogen causing feline infectious peritonitis (FIP), a lethal disease in cats, necessitating accurate diagnostic methods. This study developed and compared novel primers targeting the FCoV membrane (M) gene for enhanced detection. Specific primers were designed for the M gene and their performance evaluated using reverse transcription-PCR (RT-PCR), nested RT-PCR, and reverse transcription-quantitative PCR (RT-qPCR) on 80 clinical effusion samples from cats suspected of FIP. Specificity of assays was tested against other feline viruses, with sensitivity being assessed via serial dilutions of FCoV RNA. RT-qPCR had the highest sensitivity, detecting 9.14 × 101 copies/µL, identifying 93.75% of positive samples, followed by nested RT-PCR (87.50%, 9.14 × 104 copies/µL) and RT-PCR (61.25%, 9.14 × 106 copies/µL). All assays had 100% specificity, with no cross-reactivity to other viruses. The nested RT-PCR and RT-qPCR outperformed RT-PCR significantly, with comparable diagnostic accuracy. The novel primers targeting the FCoV M gene, coupled with RT-qPCR, delivered unparalleled sensitivity and robust reliability for detecting FCoV in clinical settings. Nested RT-PCR was equally precise and amplified diagnostic confidence with its high performance. These cutting-edge assays should revolutionize FCoV detection, offering trusted tools that seamlessly integrate into veterinary practice, empowering clinicians to manage feline infectious peritonitis with unprecedented accuracy and speed.
Keywords: FIP; RT-PCR; RT-qPCR; feline coronavirus; nested RT-PCR.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
A colloidal gold immunochromatographic test strip based on mAbs anti-N protein to detect feline coronavirus.Microbiol Spectr. 2025 Jul;13(7):e0183024. doi: 10.1128/spectrum.01830-24. Epub 2025 Jun 2. Microbiol Spectr. 2025. PMID: 40454842 Free PMC article.
-
Colorimetric Reverse Transcription Loop-Mediated Isothermal Amplification with Xylenol Orange Targeting Nucleocapsid Gene for Detection of Feline Coronavirus Infection.Viruses. 2025 Mar 14;17(3):418. doi: 10.3390/v17030418. Viruses. 2025. PMID: 40143345 Free PMC article.
-
Detection of Feline Coronavirus RNA in Cats with Feline Infectious Peritonitis and Their Housemates.Viruses. 2025 Jul 4;17(7):948. doi: 10.3390/v17070948. Viruses. 2025. PMID: 40733565 Free PMC article.
-
Laboratory-based molecular test alternatives to RT-PCR for the diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Oct 14;10(10):CD015618. doi: 10.1002/14651858.CD015618. Cochrane Database Syst Rev. 2024. PMID: 39400904
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Barker E.N., Tasker S. Advances in Molecular Diagnostics and Treatment of Feline Infectious Peritonitis. Adv. Small Anim. Care. 2020;1:161–188. doi: 10.1016/j.yasa.2020.07.011. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

