Csn5 Depletion Reverses Mitochondrial Defects in GCN5-Null Saccharomyces cerevisiae
- PMID: 40725162
- PMCID: PMC12294998
- DOI: 10.3390/ijms26146916
Csn5 Depletion Reverses Mitochondrial Defects in GCN5-Null Saccharomyces cerevisiae
Abstract
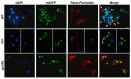
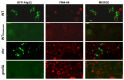
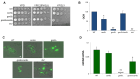
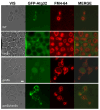
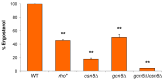
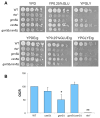
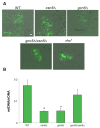
In this study, we investigated the mitochondrial defects resulting from the deletion of GCN5, a lysine-acetyltransferase, in the yeast Saccharomyces cerevisiae. Gcn5 serves as the catalytic subunit of the SAGA acetylation complex and functions as an epigenetic regulator, primarily acetylating N-terminal lysine residues on histones H2B and H3 to modulate gene expression. The loss of GCN5 leads to mitochondrial abnormalities, including defects in mitochondrial morphology, a reduced mitochondrial DNA copy number, and defective mitochondrial inheritance due to the depolarization of actin filaments. These defects collectively trigger the activation of the mitophagy pathway. Interestingly, deleting CSN5, which encodes to Csn5/Rri1 (Csn5), the catalytic subunit of the COP9 signalosome complex, rescues the mitochondrial phenotypes observed in the gcn5Δ strain. Furthermore, these defects are suppressed by exogenous ergosterol supplementation, suggesting a link between the rescue effect mediated by CSN5 deletion and the regulatory role of Csn5 in the ergosterol biosynthetic pathway.
Keywords: Saccharomyces cerevisiae; epigenetic regulation; ergosterol; lysine-acetyltransferase; mitochondria; ubiquitin–proteasome pathway.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
SAGA subunits Spt3 and Spt8 act directly and non-redundantly with TFIID in TBP recruitment in the Gcn4 transcriptome.Nucleic Acids Res. 2025 Jul 8;53(13):gkaf598. doi: 10.1093/nar/gkaf598. Nucleic Acids Res. 2025. PMID: 40637224 Free PMC article.
-
Prohibitins, Phb1 and Phb2, function as Atg8 receptors to support yeast mitophagy and also play a negative regulatory role in Atg32 processing.Autophagy. 2024 Nov;20(11):2478-2489. doi: 10.1080/15548627.2024.2371717. Epub 2024 Jul 4. Autophagy. 2024. PMID: 38964378 Free PMC article.
-
The COP9 signalosome mediates the Spt23 regulated fatty acid desaturation and ergosterol biosynthesis.FASEB J. 2020 Apr;34(4):4870-4889. doi: 10.1096/fj.201902487R. Epub 2020 Feb 19. FASEB J. 2020. PMID: 32077151
-
The emerging roles of Jab1/CSN5 in cancer.Med Oncol. 2016 Aug;33(8):90. doi: 10.1007/s12032-016-0805-1. Epub 2016 Jul 13. Med Oncol. 2016. PMID: 27412572
-
Comparison of Telomere Structure in Eukaryotes.Arch Razi Inst. 2024 Dec 31;79(6):1365-1374. doi: 10.32592/ARI.2024.79.6.1365. eCollection 2024 Dec. Arch Razi Inst. 2024. PMID: 40606259 Free PMC article. Review.
References
-
- Patel J.H., Du Y., Ard P.G., Phillips C., Carella B., Chen C.-J., Rakowski C., Chatterjee C., Lieberman P.M., Lane W.S., et al. The C-MYC Oncoprotein Is a Substrate of the Acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

