Thriving or Withering? Plant Molecular Cytogenetics in the First Quarter of the 21st Century
- PMID: 40725259
- PMCID: PMC12295485
- DOI: 10.3390/ijms26147013
Thriving or Withering? Plant Molecular Cytogenetics in the First Quarter of the 21st Century
Abstract
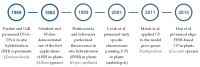
Nearly four decades have passed since fluorescence in situ hybridisation was first applied in plants to support molecular cytogenetic analyses across a wide range of species. Subsequent advances in DNA sequencing, bioinformatic analysis, and microscopy, together with the immunolocalisation of various nuclear components, have provided unprecedented insights into the cytomolecular organisation of the nuclear genome in both model and non-model plants, with crop species being perhaps the most significant. The ready availability of sequenced genomes is now facilitating the application of state-of-the-art cytomolecular techniques across diverse plant species. However, these same advances in genomics also pose a challenge to the future of plant molecular cytogenetics, as DNA sequence analysis is increasingly perceived as offering comparable insights into genome organisation. This perception persists despite the continued relevance of FISH-based approaches for the physical anchoring of genome assemblies to chromosomes. Furthermore, cytogenetic approaches cannot currently rival purely genomic methods in terms of throughput, standardisation, and automation. This review highlights the latest key topics in plant cytomolecular research, with particular emphasis on chromosome identification and karyotype evolution, chromatin and interphase nuclear organisation, chromosome structure, hybridisation and polyploidy, and cytogenetics-assisted crop improvement. In doing so, it underscores the distinctive contributions that cytogenetic techniques continue to offer in genomic research. Additionally, we critically assess future directions and emerging opportunities in the field, including those related to CRISPR/Cas-based live-cell imaging and chromosome engineering, as well as AI-assisted image analysis and karyotyping.
Keywords: FISH; chromosome; chromosome markers; cytogenetics; cytomolecular analysis; fluorescence in situ hybridisation; interphase nucleus; karyotype; plant molecular cytogenetics; plant nuclear genome.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Cancer cytogenetics in a genomics world: Wedding the old with the new.Blood Rev. 2024 Jul;66:101209. doi: 10.1016/j.blre.2024.101209. Epub 2024 May 7. Blood Rev. 2024. PMID: 38852016 Review.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
-
Integration of Newer Genomic Technologies into Clinical Cytogenetics Laboratories.Genes (Basel). 2025 Jun 4;16(6):688. doi: 10.3390/genes16060688. Genes (Basel). 2025. PMID: 40565580 Free PMC article. Review.
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
References
-
- Schwarzacher T., Heslop-Harrison J.S. Practical In Situ Hybridization. BIOS Scientific Publishers; Oxford, UK: 2000.
-
- Schubert I., Wobus U. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma. 1985;92:143–148. doi: 10.1007/BF00328466. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

