Genomic Regions Associated with Respiratory Disease in Holstein Calves in the Southern United States
- PMID: 40725399
- PMCID: PMC12294756
- DOI: 10.3390/genes16070741
Genomic Regions Associated with Respiratory Disease in Holstein Calves in the Southern United States
Abstract
Background/objectives: Bovine respiratory disease (BRD) is a common disease impacting cattle throughout the US. BRD is a multifactorial disease as disease risk varies with the genetic profile of the host, environmental conditions, and pathogen exposure. Selection for enhanced BRD resistant cattle can aid in reducing BRD. The objectives of this study were to identify loci, gene sets, and genes associated and enriched for BRD in pre- and post-weaned Holstein cattle.
Methods: Cases consisted of 2147 and 5607 calves treated for BRD as pre-weaned (0-60 days old) and post-weaned (61-420 days old) calves, respectively. Controls consisted of calves untreated for BRD that remained in the herd for 61 (n = 14,219) days for pre-weaned or 421 (n = 12,242) days for post-weaned calves. A genome-wide association analysis (GWAA) identified loci and positional candidate genes associated with BRD (uncorrected P < 1 × 10-5) for additive, dominant, and recessive inheritance models. A gene set enrichment analysis (GSEA-SNP) identified gene sets and leading-edge genes enriched (NES ≥ 3) for BRD.
Results: In pre-weaned calves, 62 loci and 123 positional candidate genes were associated (P < 1 × 10-5) in addition to the 12 gene sets and 126 leading-edge genes enriched (NES ≥ 3) for BRD. In post-weaned calves, 181 loci and 185 positional candidate genes were associated (P < 1 × 10-5), and 63 gene sets and 849 leading-edge genes were enriched (NES ≥ 3) for BRD.
Conclusions: These results provide further insight and validation of genomic regions that enhance selection for BRD resistance and for healthier cattle.
Keywords: bovine respiratory disease; cattle; gene set enrichment analysis; genome wide association analysis.
Conflict of interest statement
The authors declare no conflicts of interest, and the funders for this project had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
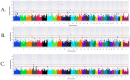
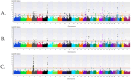
Similar articles
-
Genomic regions associated with bovine respiratory disease in pacific northwest Holstein cattle.Front Vet Sci. 2025 Jul 31;12:1637087. doi: 10.3389/fvets.2025.1637087. eCollection 2025. Front Vet Sci. 2025. PMID: 40822650 Free PMC article.
-
Heritability estimates and genome-wide association study of methane emission traits in Nellore cattle.J Anim Sci. 2024 Jan 3;102:skae182. doi: 10.1093/jas/skae182. J Anim Sci. 2024. PMID: 38967061 Free PMC article.
-
Antibiogram use on dairy cattle for bovine respiratory disease: A repeated cross-sectional study evaluating antimicrobial susceptibility of Pasteurella multocida and Mannheimia haemolytica.J Dairy Sci. 2025 Jul;108(7):7401-7414. doi: 10.3168/jds.2025-26441. Epub 2025 Apr 28. J Dairy Sci. 2025. PMID: 40306431
-
Systematic Review of the Diagnostic Accuracy of Haptoglobin, Serum Amyloid A, and Fibrinogen versus Clinical Reference Standards for the Diagnosis of Bovine Respiratory Disease.J Vet Intern Med. 2016 Jul;30(4):1356-68. doi: 10.1111/jvim.13975. Epub 2016 Jun 3. J Vet Intern Med. 2016. PMID: 27255433 Free PMC article.
-
Diagnostic accuracy of clinical illness for bovine respiratory disease (BRD) diagnosis in beef cattle placed in feedlots: A systematic literature review and hierarchical Bayesian latent-class meta-analysis.Prev Vet Med. 2016 Dec 1;135:67-73. doi: 10.1016/j.prevetmed.2016.11.006. Epub 2016 Nov 9. Prev Vet Med. 2016. PMID: 27931931
References
-
- Schaffer A.P., Larson R.L., Cernicchiaro N., Hanzlicek G.A., Bartle S.J., Thomson D.U. The association between calfhood bovine respiratory disease complex and subsequent departure from the herd, milk production, and reproduction in dairy cattle. J. Am. Vet. Med. Assoc. 2016;248:1157–1164. doi: 10.2460/javma.248.10.1157. - DOI - PubMed
-
- National Animal Health Monitoring System United States Department of Agriculture. Health and Management Practices on U.S. Dairy Operations, 2014. Report 3. [(accessed on 21 April 2025)];2018 Available online: https://www.aphis.usda.gov/sites/default/files/dairy14_dr_partiii.pdf.
-
- Urie N.J., Lombard J.E., Shivley C.B., Kopral C.A., Adams A.E., Earleywine T.J., Olson J.D., Garry F.B. Preweaned heifer management on US dairy operations: Part V. factors associated with morbidity and mortality in preweaned dairy heifer calves. J. Dairy Sci. 2018;101:9229–9244. doi: 10.3168/jds.2017-14019. - DOI - PMC - PubMed
-
- Klima C.L., Zaheer R., Cook S.R., Booker C.W., Hendrick S., Alexander T.W., McAllister T.A. Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J. Clin. Microbiol. 2014;52:438–448. doi: 10.1128/JCM.02485-13. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

