Differential Expression of Epstein-Barr Virus Sequences in Various Breast Cancer Subtypes
- PMID: 40725412
- PMCID: PMC12294798
- DOI: 10.3390/genes16070756
Differential Expression of Epstein-Barr Virus Sequences in Various Breast Cancer Subtypes
Abstract
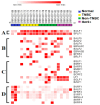
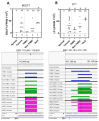
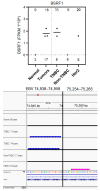
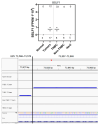
Background/Objectives: Breast cancer (BC) is the most common source of new cancer diagnoses among women and the second leading cause of cancer-related deaths in this group. The role of viral factors in the etiology, heterogeneity, and pathogenesis of this disease and its subtypes has not been incontrovertibly determined. Thus, in this study we began to address this problem by testing the hypothesis that the oncogenic Epstein-Barr virus (EBV) plays a role in this process. The approach involved determining the differential expression and predicted role of EBV gene sequences present in various subtypes of breast tumors as compared to those in control normal tissues. Methods: We utilized existing deep sequencing RNA-seq datasets derived from seventeen breast tumors and three control normal breast tissue samples to investigate the differential expression of EBV gene sequences. Results: We report three-fold higher levels of normalized total EBV-expressed sequences in tumors as compared to in control breast tissue. We also demonstrate differential expression of EBV gene transcript sequences in four categories of 26 known genes in breast cancer tumors as compared to that in normal breast tissue controls. Tumor-specific expression of EBV gene transcript sequences localized to seventeen genes; of these, tumor-specific EBV gene transcript-expressed sequences localizing to nine genes were strongly differentially expressed in a breast cancer subtype-specific manner. Furthermore, in a proof-of-concept investigation, we report, for the first time, that functional analysis of the differentially expressed integrated EBV transcript sequences demonstrate the capacity of these sequences to generate novel EBV miRNAs. We conclude that these integrated EBV sequences could potentially play a role in the pathogenesis of BC and its most aggressive subtypes. The functional role of these findings is currently under study.
Keywords: Epstein–Barr virus; HER2+ breast cancer; RNA-seq; bioinformatics; breast cancer; computational genomics; gene expression analysis; human gammaherpesvirus 4; triple-negative breast cancer; viral miRNA; viral oncogenesis.
Conflict of interest statement
The authors declare no conflicts of interest.
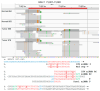
Figures





Similar articles
-
EBV-miR-BART5-5p regulates RORA to promote proliferation and migration of gastric cancer cells.PLoS One. 2025 Jul 10;20(7):e0327323. doi: 10.1371/journal.pone.0327323. eCollection 2025. PLoS One. 2025. PMID: 40638694 Free PMC article.
-
Profiling miRNA changes in Epstein-Barr virus lytic infection identifies a function for BZLF1 in upregulating miRNAs from the DLK1-DIO3 locus.PLoS Pathog. 2025 Jul 17;21(7):e1013347. doi: 10.1371/journal.ppat.1013347. eCollection 2025 Jul. PLoS Pathog. 2025. PMID: 40674434 Free PMC article.
-
The tumor immune composition of mismatch repair deficient and Epstein-Barr virus-positive gastric cancer: A systematic review.Cancer Treat Rev. 2024 Jun;127:102737. doi: 10.1016/j.ctrv.2024.102737. Epub 2024 Apr 20. Cancer Treat Rev. 2024. PMID: 38669788
-
Assessing the fitness of Epstein-Barr virus following its reactivation.J Virol. 2025 Jul 22;99(7):e0062625. doi: 10.1128/jvi.00626-25. Epub 2025 May 30. J Virol. 2025. PMID: 40444990 Free PMC article.
-
The Epstein Barr virus DNA levels as a tumor marker in EBV-associated cancers.J Cancer Res Clin Oncol. 2007 Nov;133(11):809-15. doi: 10.1007/s00432-007-0281-2. Epub 2007 Jul 25. J Cancer Res Clin Oncol. 2007. PMID: 17653573 Free PMC article. Review.
References
-
- Zhang Y., Ji Y., Liu S., Li J., Wu J., Jin Q., Liu X., Duan H., Feng Z., Liu Y., et al. Global burden of female breast cancer: New estimates in 2022, temporal trend and future projections up to 2050 based on the latest release from GLOBOCAN. J. Natl. Cancer Cent. 2025;5:287–296. doi: 10.1016/j.jncc.2025.02.002. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

