Research on Key Genes for Flowering of Bambusaoldhamii Under Introduced Cultivation Conditions
- PMID: 40725467
- PMCID: PMC12294170
- DOI: 10.3390/genes16070811
Research on Key Genes for Flowering of Bambusaoldhamii Under Introduced Cultivation Conditions
Abstract
Background: Bambusaoldhamii is an important economic bamboo species. However, flowering occurred after its introduction and cultivation, resulting in damage to the economy of bamboo forests. Currently, the molecular mechanism of flowering induced by introduction stress is still unclear. This study systematically explored the key genes and regulatory pathways of flowering in Bambusaoldhamii under introduction stress through field experiments combined with transcriptome sequencing and weighted gene co-expression network analysis (WGCNA), with the aim of providing a basis for flower-resistant cultivation and molecular breeding of bamboo.
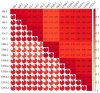
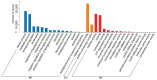
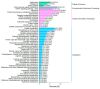
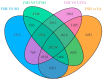
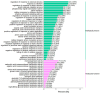
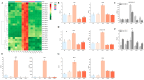
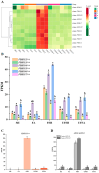
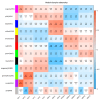
Results: The study conducted transcriptome sequencing on flowering and non-flowering Bambusaoldhamii bamboo introduced from Youxi, Fujian Province for 2 years, constructed a reference transcriptome containing 213,747 Unigenes, and screened out 36,800-42,980 significantly differentially expressed genes (FDR < 0.05). The results indicated that the photosensitive gene CRY and the temperature response gene COR413-PM were significantly upregulated in the flowering group; the expression level of the heavy metal detoxification gene MT3 increased by 27.77 times, combined with the upregulation of the symbiotic signaling gene NIN. WGCNA analysis showed that the expression level of the flower meristem determination gene AP1/CAL/FUL in the flowering group was 90.38 times that of the control group. Moreover, its expression is regulated by the cascade synergy of CRY-HRE/RAP2-12-COR413-PM signals.
Conclusions: This study clarifies for the first time that the stress of introducing Bambusaoldhamii species activates the triad pathways of photo-temperature signal perception (CRY/COR413-PM), heavy metal detoxification (MT3), and symbiotic regulation (NIN), collaboratively driving the AP1/CAL/FUL gene expression network and ultimately triggering the flowering process.
Keywords: Bambusaoldhamii; environmental stresses; flowering; introduced cultivation; key genes.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Transcriptomic Identification of Key Genes Responding to High Heat Stress in Moso Bamboo (Phyllostachys edulis).Genes (Basel). 2025 Jul 23;16(8):855. doi: 10.3390/genes16080855. Genes (Basel). 2025. PMID: 40869903 Free PMC article.
-
Comparative transcriptome analysis reveals the potential mechanism of seed germination promoted by trametenolic acid in Gastrodia elata Blume.Sci Rep. 2025 Jul 24;15(1):26869. doi: 10.1038/s41598-025-12269-z. Sci Rep. 2025. PMID: 40702084 Free PMC article.
-
AdBSK1-Mediated Hormone Signaling Regulates Flowering Transition in Actinidia deliciosa 'Guichang'.Genes (Basel). 2025 Jun 28;16(7):760. doi: 10.3390/genes16070760. Genes (Basel). 2025. PMID: 40725416 Free PMC article.
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- de Jesus D.A., Batista D.M., Monteiro E.F., Salzman S., Carvalho L.M., Santana K., André T. Structural changes and adaptative evolutionary constraints in FLOWERING LOCUS T and TERMINAL FLOWER1-like genes of flowering plants. Front. Genet. 2022;13:954015. doi: 10.3389/fgene.2022.954015. - DOI - PMC - PubMed
-
- Liu J., Wu Y., Zhou L., Zhang A., Wang S., Liu Y., Yang D., Wang S. Influence of flowering on the anatomical structure, chemical components and carbohydrate metabolism of Bambusa tuldoides culms at different ages. Front. Plant Sci. 2023;14:1260302. doi: 10.3389/fpls.2023.1260302. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

