Integrating Imaging and Genomics in Amelogenesis Imperfecta: A Novel Diagnostic Approach
- PMID: 40725478
- PMCID: PMC12294184
- DOI: 10.3390/genes16070822
Integrating Imaging and Genomics in Amelogenesis Imperfecta: A Novel Diagnostic Approach
Abstract
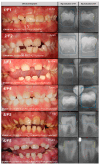
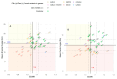
Background/Objectives: Amelogenesis imperfecta (AI) represents a heterogeneous group of inherited disorders affecting the quality and quantity of dental enamel, making clinical diagnosis challenging. This study aimed to identify genetic variants in Slovenian patients with non-syndromic AI and to evaluate enamel morphology using radiographic parameters. Methods: Whole exome sequencing (WES) was performed on 24 AI patients and their families. Panoramic radiographs (OPTs) were analyzed using Fiji ImageJ to assess crown dimensions, enamel angle (EA), dentine angle (DA), and enamel-dentine mineralization ratio (EDMR) in lower second molar buds, compared to matched controls (n = 24). Two observers independently assessed measurements, and non-parametric tests compared EA, DA, and EDMR in patients with and without disease-causing variants (DCVs). Statistical models, including bootstrap-validated random forest and logistic regression, assessed variable influences. Results: DCVs were identified in ENAM (40% of families), AMELX (15%), and MMP20 (10%), including four novel variants. AI patients showed significant enamel deviations with high reproducibility, particularly in hypomineralized and hypoplastic regions. DA and EDMR showed significant correlations with DCVs (p < 0.01). A bootstrap-validated random forest model yielded a 90% (84.0-98.0%) AUC-estimated predictive power. Conclusions: These findings highlight a novel and reproducible radiographic approach for detecting developmental enamel defects in AI and support its diagnostic potential.
Keywords: amelogenesis imperfecta; disease-causing variants; imaging genomics; molecular genetics; panoramic; radiography.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





Similar articles
-
Genetic Screening of a Nonsyndromic Amelogenesis Imperfecta Patient Cohort Using a Custom smMIP Reagent for Selective Enrichment of Target Loci.Hum Mutat. 2025 Jul 22;2025:8942542. doi: 10.1155/humu/8942542. eCollection 2025. Hum Mutat. 2025. PMID: 40741335 Free PMC article.
-
Fractal analysis of mandibular bone structure in children with amelogenesis imperfecta.Bone. 2025 Oct;199:117571. doi: 10.1016/j.bone.2025.117571. Epub 2025 Jun 28. Bone. 2025. PMID: 40582393
-
Human and mouse enamel phenotypes resulting from mutation or altered expression of AMEL, ENAM, MMP20 and KLK4.Cells Tissues Organs. 2009;189(1-4):224-9. doi: 10.1159/000151378. Epub 2008 Aug 19. Cells Tissues Organs. 2009. PMID: 18714142 Free PMC article.
-
Ameloblastin and its multifunctionality in amelogenesis: A review.Matrix Biol. 2024 Aug;131:62-76. doi: 10.1016/j.matbio.2024.05.007. Epub 2024 May 28. Matrix Biol. 2024. PMID: 38815936 Free PMC article. Review.
-
Dental treatment approaches of amelogenesis imperfecta in children and young adults: A systematic review of the literature.J Esthet Restor Dent. 2024 Jun;36(6):881-891. doi: 10.1111/jerd.13191. Epub 2024 Jan 23. J Esthet Restor Dent. 2024. PMID: 38258433
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

