DCAF16-Based Covalent Degradative Handles for the Modular Design of Degraders
- PMID: 40726802
- PMCID: PMC12291113
- DOI: 10.1021/acscentsci.5c00959
DCAF16-Based Covalent Degradative Handles for the Modular Design of Degraders
Abstract
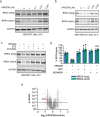
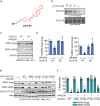
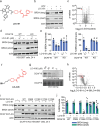
While targeted protein degradation is a powerful strategy for eliminating disease-causing proteins, the rational design of monovalent or molecular glue degraders remains challenging. In this study, we generated a library of BET-domain inhibitor JQ1 analogs bearing elaborated electrophilic handles to identify permissive covalent degradative handles and E3 ligase pairs. We identified an elaborated fumaramide handle that, when appended onto JQ1, led to the proteasome-dependent degradation of BRD4. We revealed that the E3 ubiquitin ligase CUL4DCAF16a common E3 ligase target of electrophilic degraderswas responsible for BRD4 loss by covalently targeting C173 on DCAF16. While this original fumaramide handle was not permissive to the degradation of other neo-substrates, a truncated version of this handle attached to JQ1 was still capable of degrading BRD4, now through targeting both C173 and C178. This truncated fumaramide handle, when appended to various protein targeting ligands, was also more permissive in degrading other neo-substrates, including CDK4/6, SMARCA2/4, the androgen receptor (AR), as well as the undruggable AR truncation variant AR-V7. We have identified a unique DCAF16-targeting covalent degradative handle that can be transplanted across several protein-targeting ligands to induce the degradation of their respective targets for the modular design of monovalent or bifunctional degraders.
© 2025 The Authors. Published by American Chemical Society.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
