Zanthoxylum schinifolium extracts enhance 3T3-L1 adipocyte differentiation via CHOP inhibition and PPARγ activation
- PMID: 40726805
- PMCID: PMC12302382
- DOI: 10.1080/19768354.2025.2536022
Zanthoxylum schinifolium extracts enhance 3T3-L1 adipocyte differentiation via CHOP inhibition and PPARγ activation
Abstract
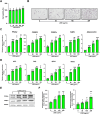
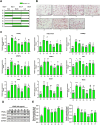
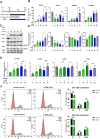
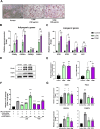
Zanthoxylum schinifolium (ZS), which is widely used as a seasoning and traditional medicine in East Asia, has demonstrated pharmacological potential. This study investigated the effects of the leaf and twig extracts of ZS (LZSE and TZSE, respectively), which are native to the Honam region of Korea, on adipocyte differentiation and assessed the ligand-binding energy score of their components to bind peroxisome proliferator-activated receptor gamma (PPARγ), a critical regulator of adipogenesis and metabolic health. LZSE and TZSE were prepared using 70% ethanol, and their molecular effects on adipocyte differentiation were evaluated in 3T3-L1 preadipocytes. Single compounds from the extracts were identified using UPLC-ESI-Q-TOF-MS, and their ligand-binding energy scores were calculated via in silico molecular docking studies. PPARγ activity was further confirmed through reporter assays. LZSE and TZSE significantly promoted adipocyte differentiation, as demonstrated by morphological changes and the increased mRNA and protein levels of key adipogenic and lipogenic genes, such as PPARγ and CCAAT-enhancer-binding protein alpha (C/EBPα). LZSE specifically enhanced adipogenesis without inducing cytotoxicity, attributed to the inhibition of C/EBP homologous protein (CHOP) and stimulation of mitotic expansion. Additionally, UPLC-ESI-Q-TOF-MS identified several active compounds in LZSE and TZSE, and in silico docking revealed the high binding affinity of these compounds for the full-agonist ligand-binding domain of PPARγ. LZSE and TZSE could emerge as novel antidiabetic drug candidates based on their effects on adipocyte differentiation and PPARγ activation. Furthermore, the active compounds identified in these extracts hold promise as tentative PPARγ agonists, highlighting their therapeutic potential in the treatment of metabolic disorders.
Keywords: Adipocytes; Zanthoxylum schinifolium; adipogenesis; differentiation; molecular docking.
© 2025 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


Similar articles
-
Melatonin and Its Metabolites Can Serve as Agonists on the Aryl Hydrocarbon Receptor and Peroxisome Proliferator-Activated Receptor Gamma.Int J Mol Sci. 2023 Oct 23;24(20):15496. doi: 10.3390/ijms242015496. Int J Mol Sci. 2023. PMID: 37895177 Free PMC article.
-
Cucurbitacin E Suppresses Adipogenesis and Lipid Accumulation in 3T3-L1 Adipocytes Without Cytotoxicity.Biomedicines. 2025 Jul 25;13(8):1826. doi: 10.3390/biomedicines13081826. Biomedicines. 2025. PMID: 40868081 Free PMC article.
-
Bisphenol F promoted the differentiation of preadipocytes via ER-mediated PI3K/AKT pathway.Food Chem Toxicol. 2025 Aug 5;205:115678. doi: 10.1016/j.fct.2025.115678. Online ahead of print. Food Chem Toxicol. 2025. PMID: 40752664
-
Insulin interacts with PPARγ agonists to promote bovine adipocyte differentiation.Domest Anim Endocrinol. 2024 Jul;88:106848. doi: 10.1016/j.domaniend.2024.106848. Epub 2024 Mar 29. Domest Anim Endocrinol. 2024. PMID: 38574690 Review.
-
Unraveling the rationale and conducting a comprehensive assessment of KD025 (Belumosudil) as a candidate drug for inhibiting adipogenic differentiation-a systematic review.Naunyn Schmiedebergs Arch Pharmacol. 2024 May;397(5):2681-2699. doi: 10.1007/s00210-023-02834-6. Epub 2023 Nov 15. Naunyn Schmiedebergs Arch Pharmacol. 2024. PMID: 37966572
References
LinkOut - more resources
Full Text Sources
Research Materials
