Interferon Regulatory Factor 4 dose-dependently controls peripheral Treg cell differentiation and homeostasis by modulating chromatin accessibility in mice
- PMID: 40726986
- PMCID: PMC12302998
- DOI: 10.3389/fimmu.2025.1604888
Interferon Regulatory Factor 4 dose-dependently controls peripheral Treg cell differentiation and homeostasis by modulating chromatin accessibility in mice
Abstract
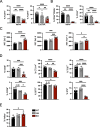
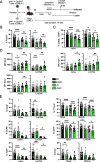
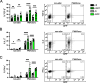
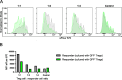
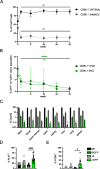
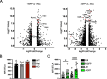
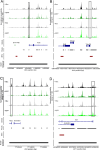
FoxP3+ regulatory T (Treg) cells restrict excessive immune responses and immunopathology as well as reactivity to self or environmental antigens and thus are crucial for peripheral immune tolerance. The transcription factor Interferon Regulatory Factor 4 (IRF4) controls differentiation and function of T cells. In Treg cells, IRF4 is required for peripheral activation and maturation to effector Treg (eTreg) cells with enhanced suppressive function. However, the mechanisms of Treg cell regulation by IRF4 are not fully understood. Here, we analyze the role of IRF4 in differentiation and maintenance of Treg cells using IRF4-deficient mice and a T cell transfer model, that allows Irf4 inactivation in peripheral T cells. We demonstrate that loss of one Irf4 allele already results in impaired eTreg cell differentiation and decreased Treg cell homeostasis, indicating that IRF4 controls peripheral Treg cell differentiation in a gene dosage dependent mode. Peripheral Irf4 inactivation was also associated with enhanced production of inflammatory but also inhibitory cytokines by Treg cells. ATAC sequencing of Treg cells after mutation of one or both Irf4 alleles revealed regions with altered accessibility in genes involved in Treg cell function. In the FoxP3 gene, Irf4 inactivation resulted in reduced ATAC signals in the promoter region and in the conserved non-coding sequence (CNS) 2, required for stability of FoxP3 expression in peripheral Treg cells in response to TCR stimulation. IRF4-deficient Treg cells also displayed a reduction in open chromatin in several Treg cell specific super enhancers, mainly located in proximity to potential IRF4 binding sites. In conclusion, our results demonstrate that IRF4 controls peripheral Treg cell differentiation and homeostasis in a gene dosage dependent manner.
Keywords: FoxP3; Interferon Regulatory Factor 4; T cells; assay for transposase-accessible chromatin (ATAC) sequencing; regulatory T cells.
Copyright © 2025 Voß, Schmidt, Harberts, Spohn, Bradtke, Borchers, Fertig, Schmid, Koch-Nolte, Krebs, Raczkowski and Mittrücker.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Comprehensive single-cell chromatin and transcriptomic profiling of peripheral immune cells in nonsegmental vitiligo.Br J Dermatol. 2025 Jun 20;193(1):115-124. doi: 10.1093/bjd/ljaf041. Br J Dermatol. 2025. PMID: 39888372
-
Attenuation of TCR-induced transcription by Bach2 controls regulatory T cell differentiation and homeostasis.Nat Commun. 2020 Jan 14;11(1):252. doi: 10.1038/s41467-019-14112-2. Nat Commun. 2020. PMID: 31937752 Free PMC article.
-
The kinase ITK controls a Ca2+-mediated switch that balances TH17 and Treg cell differentiation.Sci Signal. 2024 Jul 23;17(846):eadh2381. doi: 10.1126/scisignal.adh2381. Epub 2024 Jul 23. Sci Signal. 2024. PMID: 39042726 Free PMC article.
-
Role of the intestinal flora-immunity axis in the pathogenesis of rheumatoid arthritis-mechanisms regulating short-chain fatty acids and Th17/Treg homeostasis.Mol Biol Rep. 2025 Jun 21;52(1):617. doi: 10.1007/s11033-025-10714-w. Mol Biol Rep. 2025. PMID: 40544212 Review.
-
Microbiota-dependent modulation of intestinal anti-inflammatory CD4+ T cell responses.Semin Immunopathol. 2025 Apr 1;47(1):23. doi: 10.1007/s00281-025-01049-6. Semin Immunopathol. 2025. PMID: 40167791 Review.
References
-
- Raczkowski F, Ritter J, Heesch K, Schumacher V, Guralnik A, Höcker L, et al. The transcription factor Interferon Regulatory Factor 4 is required for the generation of protective effector CD8+ T cells. Proc Natl Acad Sci USA. (2013) 110:15019–24. doi: 10.1073/pnas.1309378110, PMID: - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

