Multi-level channel-spatial attention and light-weight scale-fusion network (MCSLF-Net): multi-level channel-spatial attention and light-weight scale-fusion transformer for 3D brain tumor segmentation
- PMID: 40727355
- PMCID: PMC12290658
- DOI: 10.21037/qims-2025-354
Multi-level channel-spatial attention and light-weight scale-fusion network (MCSLF-Net): multi-level channel-spatial attention and light-weight scale-fusion transformer for 3D brain tumor segmentation
Abstract
Background: Gliomas, the most aggressive primary tumors in the central nervous system, are characterized by high morphological heterogeneity and diffusely infiltrating boundaries. Such complexity poses significant challenges for accurate segmentation in clinical practice. Although deep learning methods have shown promising results, they often struggle to achieve a satisfactory trade-off among precise boundary delineation, robust multi-scale feature representation, and computational efficiency, particularly when processing high-resolution three-dimensional (3D) magnetic resonance imaging (MRI) data. Therefore, the aim of this study is to develop a novel 3D segmentation framework that specifically addresses these challenges, thereby improving clinical utility in brain tumor analysis. To accomplish this, we propose a multi-level channel-spatial attention and light-weight scale-fusion network (MCSLF-Net), which integrates a multi-level channel-spatial attention mechanism (MCSAM) and a light-weight scale-fusion module. By strategically enhancing subtle boundary features while maintaining a compact network design, our approach seeks to achieve high accuracy in delineating complex glioma morphologies, reduce computational burden, and provide a more clinically feasible segmentation solution.
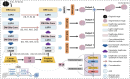
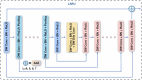
Methods: We propose MCSLF-Net, a network integrating two key components: (I) MCSAM: by strategically inserting a 3D channel-spatial attention module at critical semantic layers, the network progressively emphasizes subtle, infiltrative edges and small, easily overlooked contours. This avoids reliance on an additional edge detection branch while enabling fine-grained localization in ambiguous transitional regions. (II) Light-weight scale fusion unit (LSFU): leveraging depth-wise separable convolutions combined with multi-scale atrous (dilated) convolutions, LSFU enhances computational efficiency and adapts to varying feature requirements at different network depths. In doing so, it effectively captures small infiltrative lesions as well as extensive tumor areas. By coupling these two modules, MCSLF-Net balances global contextual information with local fine-grained features, simultaneously reducing the computational burden typically associated with 3D medical image segmentation.
Results: Extensive experiments on the BraTS 2019, BraTS 2020, and BraTS 2021 datasets validated the effectiveness of our approach. On BraTS 2021, MCSLF-Net achieved a mean Dice similarity coefficient (DSC) of 0.8974 and a mean 95th percentile Hausdorff distance (HD95) of 2.52 mm. Notably, it excels in segmenting intricate transitional areas, including the enhancing tumor (ET) region and the tumor core (TC), thereby demonstrating superior boundary delineation and multi-scale feature fusion capabilities relative to existing methods.
Conclusions: These findings underscore the clinical potential of deploying multi-level channel-spatial attention and light-weight multi-scale fusion strategies in high-precision 3D glioma segmentation. By striking an optimal balance among boundary accuracy, multi-scale feature capture, and computational efficiency, the proposed MCSLF-Net offers a practical framework for further advancements in automated brain tumor analysis and can be extended to a range of 3D medical image segmentation tasks.
Keywords: Brain tumor segmentation; light-weight scale fusion; multi-level attention mechanism; transformer-based network.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-2025-354/coif). The authors have no conflicts of interest to declare.
Figures



Similar articles
-
A novel image segmentation network with multi-scale and flow-guided attention for early screening of vaginal intraepithelial neoplasia (VAIN).Med Phys. 2025 Aug;52(8):e18041. doi: 10.1002/mp.18041. Med Phys. 2025. PMID: 40804792
-
VMDU-net: a dual encoder multi-scale fusion network for polyp segmentation with Vision Mamba and Cross-Shape Transformer integration.Front Artif Intell. 2025 Jun 18;8:1557508. doi: 10.3389/frai.2025.1557508. eCollection 2025. Front Artif Intell. 2025. PMID: 40607455 Free PMC article.
-
A novel recursive transformer-based U-Net architecture for enhanced multi-scale medical image segmentation.Comput Biol Med. 2025 Sep;196(Pt A):110658. doi: 10.1016/j.compbiomed.2025.110658. Epub 2025 Jul 6. Comput Biol Med. 2025. PMID: 40618700
-
Magnetic resonance perfusion for differentiating low-grade from high-grade gliomas at first presentation.Cochrane Database Syst Rev. 2018 Jan 22;1(1):CD011551. doi: 10.1002/14651858.CD011551.pub2. Cochrane Database Syst Rev. 2018. PMID: 29357120 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
-
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131:803-20. 10.1007/s00401-016-1545-1 - DOI - PubMed
-
- Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, Shinohara RT, Berger C, Ha SM, Rozycki M. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv:1811.02629 [Preprint]. 2018. Available online: https://arxiv.org/abs/1811.02629
-
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18. Cham: Springer; 2015:234-41.
LinkOut - more resources
Full Text Sources
