Identification of transcriptomic sepsis endotypes in sub-Saharan Africa: derivation, validation, and global alignment in two Ugandan cohorts
- PMID: 40728637
- PMCID: PMC12405036
- DOI: 10.1007/s00134-025-08047-0
Identification of transcriptomic sepsis endotypes in sub-Saharan Africa: derivation, validation, and global alignment in two Ugandan cohorts
Abstract
Purpose: Sub-Saharan Africa carries the highest global burden of critical illness, yet transcriptomic sepsis endotypes have not been defined in the region. Their clinical relevance and alignment with endotypes identified in high-income countries (HICs) remain unknown.
Methods: We analyzed data from two prospective observational cohorts of critically ill adults with sepsis in Uganda (discovery cohort [Tororo, rural], N = 243; validation cohort [Entebbe, urban], N = 112). Unsupervised clustering of whole-blood RNAseq data was used to identify endotypes in the discovery cohort. A random forest classifier was used to predict endotype assignment in the validation cohort. Differential gene expression, pathway enrichment, and digital cytometry were used to define endotype pathobiology and determine overlap with HIC-derived endotypes.
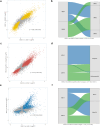
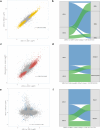
Results: Two endotypes-Uganda Sepsis Endotypes 1 (USE-1) and 2 (USE-2)-were identified in the discovery cohort. USE-2, marked by neutrophil-driven innate immune activation and lymphocyte suppression, was associated with greater physiological severity and higher mortality (41.3% vs. 22.0%; absolute difference 19.3%, 95% CI 7.6-30.9%), irrespective of HIV, tuberculosis, or malaria infection. A 13-gene classifier (misclassification rate 1.43%) replicated two endotypes in the validation cohort with similar biological and clinical profiles. USE-2 showed strong transcriptional overlap with SRS1 and inflammopathic endotypes but only modest concordance in patient-level assignments. Overlap with Mars1 was variable.
Conclusions: We identified two transcriptomic sepsis endotypes in Uganda that reflect inter-individual differences in targetable pathobiology and confer prognostic enrichment across high-burden infections. Divergence from HIC-derived endotypes highlights the need for sepsis classifications that are both globally relevant and locally responsive.
Keywords: Africa; HIV; High-throughput nucleotide sequencing; Precision medicine; Sepsis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflicts of interest: MJC reports consulting fees from Vertex Pharmaceuticals and Veracyte unrelated to the submitted work. The remaining authors declare no conflicts of interest. Ethical approval: Each enrolled participant or their surrogate provided written informed consent. Study protocols were approved by ethics committees at Columbia University (AAAR1450), Uganda Virus Research Institute (GC/127/17/02–06/582), and Uganda National Council for Science and Technology (HS2308). AI declaration: ChatGPT Education-4o (OpenAI) was used for grammatical correction and to enhance manuscript readability.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

