Molecular Interactions Between NK Cells and Acute Leukemic Cells: KIR2DL5 Drastically Limits NK Cell Responses
- PMID: 40728766
- PMCID: PMC12307528
- DOI: 10.1007/s10875-025-01913-y
Molecular Interactions Between NK Cells and Acute Leukemic Cells: KIR2DL5 Drastically Limits NK Cell Responses
Abstract
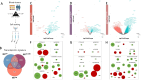
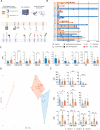
Natural Killer (NK) cells naturally recognize and eliminate leukemic cells. However, the molecular interactions that govern these responses are diverse due to the large number of activating and inhibitory NK receptors that modulate NK functions and the diversity of corresponding ligands that are differentially expressed in acute lymphoblastic and myeloblastic leukemias. We identified resting NKG2A+ NK cells and NKG2A+KIR+ NK cell subsets as the most effective in eliminating lymphoid and myeloid leukemic cells respectively. The NKG2A+KIR±CD57- cell subsets show high expression of activating receptors and a functional transcriptomic profile, but differ in KIR2DL5 expression. The frequency of KIR2DL5+ NK cells increases with the number of expressed KIR. Furthermore, KIR2DL5 is preferentially co-expressed with KIR2DL1 and is negatively regulated by NKG2A. Of note, CD57 expression, regardless of the NK cell subset considered, is associated with reduced receptor expression, consistent with its reduced cytotoxic potential. Furthermore, molecular interactions between NK cells and leukemic cells influence NK cell responses, particularly the inhibitory KIR2DL5-PVR axis. The integration of these data is of importance for the optimization of NK cell-based immunotherapies, as the selection of NK cell donors represents a key parameter for the improvement of these therapies.
Keywords: Acute leukemia; Heterogeneity; NK cells.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interest: The authors declare no competing interests.
Figures



References
-
- Herberman RB, Nunn ME, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. I. Distribution of reactivity and specificity. Int J Cancer. 1975;16(2):216–29. - PubMed
-
- Anfossi N, André P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–42. - PubMed
-
- Bottino C, Castriconi R, Moretta L, Moretta A. Cellular ligands of activating NK receptors. Trends Immunol. 2005;26:221–6. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

