Gene expression and alternative splicing reveal the co-regulation of host response mechanisms to avian leukosis virus subgroup J-infected in laying hens
- PMID: 40729809
- PMCID: PMC12329114
- DOI: 10.1016/j.psj.2025.105554
Gene expression and alternative splicing reveal the co-regulation of host response mechanisms to avian leukosis virus subgroup J-infected in laying hens
Abstract
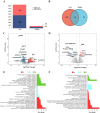
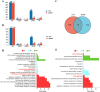
Avian leukosis in China has spread from broiler chickens to the local breeds and commercial laying hens. Studying resistance to avian leukosis is important for disease-resistant breeding programs. Gene expression and different transcripts may affect immune function. In this study, we compared five naturally infected Rhode Island Red (RIR) hens carrying tumor with five uninfected individuals to explore avian leukosis virus subgroup J (ALV-J) induced differences in gene expression and alternative splicing (AS) in the liver, spleen caused. Analyses revealed 847, 80 differentially expressed genes (DEGs), along with 207, 167 differential alternative splicing genes (DASGs) in the liver, spleen respectively. Most differential splicing events involved exon skipping. Although most genes showed no significant expression changes, their protein spatial structures were altered by AS. In the liver, microtubule cytoskeleton-related functions were co-regulated by both gene expression and splicing, with CCSER2 and MAPT exhibiting the highest splicing frequency. In the spleen, splicing predominantly affected RNA-processing genes, where PKLR and SRSF7 functioned as key regulators. Notably, PKLR-interacting genes (THRSP, ADH1C, AQP3) were significantly downregulated in infected groups, potentially promoting viral replication and tumor proliferation. These findings demonstrate that AS contributes to the host response to ALV-J infection through multiple mechanisms, including protein structural remodeling and dysregulation of coordinated interaction networks. This study provides new insights into the genetic basis of ALV-J resistance in laying hens.
Keywords: Alternative splicing; Avian leukosis virus subgroup J; Gene expression; Laying hen.
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Disclosures The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

