Comparing non-machine learning vs. machine learning methods for Ki67 scoring in gastrointestinal neuroendocrine tumors
- PMID: 40730596
- PMCID: PMC12307702
- DOI: 10.1038/s41598-025-08778-6
Comparing non-machine learning vs. machine learning methods for Ki67 scoring in gastrointestinal neuroendocrine tumors
Abstract
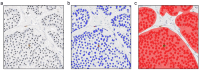
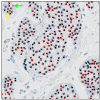
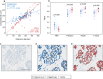
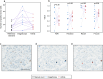
The Ki67 score is a crucial prognostic biomarker for neuroendocrine tumors, but its manual assessment is labor-intensive, requiring the counting of 500-2,000 cells in hotspots. Digital image analysis could streamline this process, yet few comprehensive comparisons exist between different tools. We compared a non-machine learning (non-ML) tool (ImageScope, Leica Biosystems) with a machine learning (ML) tool (Aiforia Create, Aiforia Technologies) on Ki67-stained slides from 10 low proliferative neuroendocrine tumor cases (Ki67 score < 5%, eight regions per slide). Performance metrics based on the coordinates of detected cells were used to assess the capability of image analysis tools to detect (i) total and (ii) Ki67 positive tumor cells, and consequently calculate the (iii) Ki67 score. Manual scoring by an experienced pathologist was used as the reference standard. The ML compared to the non-ML tool showed better performance metrics (F-score 0.90 vs. 0.74) in detecting the tumor cells. Also, the ML tool had a higher agreement with the reference standard in detecting tumor cells (ICC 0.91 vs. 0.62), Ki67 positive tumor cells (ICC 0.70 vs. 0.24), and the Ki67 score (ICC 0.86 vs. 0.45). Our findings highlight the enhanced accuracy of ML-based image analysis in detecting the correct tumor cells, outperforming traditional methods.
Keywords: Image analysis; Immunohistochemistry; Intra-observer agreement; Ki67; Machine learning; Neuroendocrine tumor (NET).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

References
-
- Rindi, G. et al. A common classification framework for neuroendocrine neoplasms: an international agency for research on cancer (IARC) and world health organization (WHO) expert consensus proposal. Mod. Pathology: Offi. J. United States Can. Acad. Pathol. Inc31, 1770–1786. 10.1038/s41379-018-0110-y (2018). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

