Autocrine interferon poisoning mediates ADAR1-dependent synthetic lethality in BRCA1/2-mutant cancers
- PMID: 40730818
- PMCID: PMC12307730
- DOI: 10.1038/s41467-025-62309-5
Autocrine interferon poisoning mediates ADAR1-dependent synthetic lethality in BRCA1/2-mutant cancers
Abstract
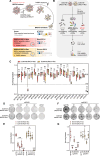
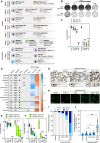
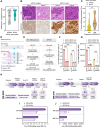
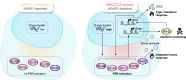
ADAR1 is an RNA editing enzyme which prevents autoimmunity by blocking interferon responses triggered by cytosolic RNA sensors, and is a potential target in immuno-oncology. However, predictive biomarkers for ADAR1 inhibition are lacking. Using multiple in vitro and in vivo systems, we show that BRCA1/2 and ADAR1 are synthetically lethal, and that ADAR1 activity is upregulated in BRCA1/2-mutant cancers. ADAR1 depletion in BRCA1-mutant cells causes an increase in R-loops and consequently, an upregulation of cytosolic nucleic acid sensing pattern recognition receptors (PRR), events which are associated with a tumor cell-autonomous type I interferon and integrated stress response. This ultimately causes autocrine interferon poisoning. Consistent with a key role of R-loops in this process, exogenous RNase H1 expression reverses the synthetic lethality. Pharmacological suppression of cell-autonomous interferon responses or transcriptional silencing of cytosolic nucleic acid sensing PRR are also sufficient to abrogate ADAR1 dependency in BRCA1-mutant cells, in line with autocrine interferon poisoning playing a central part in this synthetic lethality. Our findings provide a preclinical rationale for assessing ADAR1-targeting agents in BRCA1/2-mutant cancers, and introduces a conceptually novel approach to synthetic lethal treatments, which exploits tumor cell-intrinsic cytosolic immunity as a targetable vulnerability of cancer cells.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: R.M.C. makes the following disclosures: is named inventor on a patent describing the use of ADAR1 inhibitors as anti-cancer agents (WO/2024/189433). J.Z. makes the following disclosures: has received research funding and travel subsidies from Menarini. B.P. makes the following disclosures: has received research funding as part of an institutional program from: Astra Zeneca, Daiichi Sankyo, Gilead, Seagen, MSD; receives and/or has received consulting fees from: Astra Zeneca (institutional), Seagen (institutional), Gilead (institutional), Novartis (institutional), Lilly (institutional), MSD (institutional), Pierre Fabre (personal), Daiichi Sankyo (institutional/personal), Olema (institutional); has received travel subsidies from: Astra Zeneca; Pierre Fabre; MSD; Daiichi Sankyo, Pfizer. A.N.J.T. makes the following disclosures: Receipt of grants / research support: AstraZeneca research costs associated with OlympiA, Myriad Genetics research support for TNT Trial, Breast Cancer Now research grant to Institution, CRUK research grant to Institution. Receipt of honoraria or consultation fees: AstraZeneca, Inbiomotion, Gilead, Innovation in Breast Cancer Symposium, SABCS, GBCC, Cancer Panel, Research to Practise, AACR, Aicme, Penn Medicine, PAGE Therapeutics, Ellipses, VHIO, Dana Faber David Livingston Memorial Symposium, Tango Therapuetics, Guardant, CRUK, Neogenomics, Merck. Educational seminar: Guardant. Stock option: InBioMotion. Royalties: Benefits from ICR’s Inventors Scheme associated with patents for use of several PARP inhibitors in DNA repair-deficient cancers held by AstraZeneca with royalty payments to A.N.J.T.’s personal account and research accounts at the Institute of Cancer Research, and to the Institute of Cancer Research. Institutional financial interests: Royalties as above from AstraZeneca. Research costs from AstraZeneca, Myriad Genetics. Non-financial interests: Leadership/Guideline Advisor roles: Director of Breast Cancer Now Research Centre—ICR/KCL, St Gallen Early Breast Cancer Guidelines Panel, ESMO Early Breast Cancer Guidelines Committee, ESMO Scientific Committee, Chair—CRUK Clinical Research Committee, BCRF SAB member, Strategy Committee UK NCRI, Trustee The Cridlan Ross Smith Charitable Trust. C.J.L. makes the following disclosures: receives and/or has received research funding from: AstraZeneca, Merck KGaA, Artios, Neophore; has received consultancy, SAB membership or honoraria payments from: FoRx, Syncona, Sun Pharma, Gerson Lehrman Group, Merck KGaA, Vertex, AstraZeneca, Tango Therapeutics, 3rd Rock, Ono Pharma, Artios, Abingworth, Tesselate, Dark Blue Therapeutics, Pontifax, Astex, Neophore, Glaxo Smith Kline, Dawn Bioventures, Blacksmith Medicines, ForEx; has stock in: Tango, Ovibio, Hysplex, Tesselate, Ariceum. C.J.L. is also named inventor on patents describing the use of DNA repair inhibitors and stands to gain from their development and use as part of the ICR “Rewards to Inventors” scheme and also reports benefits from this scheme associated with patents for PARP inhibitors paid into C.J.L.’s personal accounts and research accounts at the Institute of Cancer Research. C.J.L. is also named inventor on a patent describing the use of ADAR1 inhibitors as anti-cancer agents (WO 2024/189433 A1). S.P.V. makes the following disclosures: has received research funding from Hoffman La Roche and AstraZeneca for unrelated research projects. As part of the Drug Development Department (DITEP), S.P.V. is principal investigator or sub-investigator of clinical trials from Abbvie, Agios Pharmaceuticals, Amgen, Argen-X Bvba, Arno Therapeutics, Astex Pharmaceuticals, Astra Zeneca, Aveo, Bayer Healthcare Ag, Bbb Technologies Bv, Blueprint Medicines, Boehringer Ingelheim, Bristol Myers Squibb, Celgene Corporation, Chugai Pharmaceutical Co., Clovis Oncology, Daiichi Sankyo, Debiopharm S.A., Eisai, Eli Lilly, Exelixis, Forma, Gamamabs, Genentech, Inc., GlaxoSmithKline, H3 Biomedicine, Inc, Hoffmann La Roche Ag, Innate Pharma, Iris Servier, Janssen Cilag, Kyowa Kirin Pharm. Dev., Inc., Loxo Oncology, Lytix Biopharma As, Medimmune, Menarini Ricerche, Merck Sharp & Dohme Chibret, Merrimack Pharmaceuticals, Merus, Millennium Pharmaceuticals, Nanobiotix, Nektar Therapeutics, Novartis Pharma, Octimet Oncology Nv, Oncoethix, Onyx Therapeutics, Orion Pharma, Oryzon Genomics, Pfizer, Pharma Mar, Pierre Fabre, Roche, Sanofi Aventis, Taiho Pharma, Tesaro Inc, and Xencor. S.P.V. has participated to advisory boards for Merck KGaA. S.P.V. is also named inventor on a patent describing the use of ADAR1 inhibitors as anti-cancer agents (WO 2024/189433 A1). All other authors have no conflicts of interest or financial interests to disclose.
Figures



References
MeSH terms
Substances
Grants and funding
- 2023-165-PLBIO23-139/Institut National Du Cancer (French National Cancer Institute)
- 2023-165-PLBIO23-139/Institut National Du Cancer (French National Cancer Institute)
- ARCPGA2023010005866_6378/Fondation ARC pour la Recherche sur le Cancer (ARC Foundation for Cancer Research)
- ARCPGA2023010005866_6378/Fondation ARC pour la Recherche sur le Cancer (ARC Foundation for Cancer Research)
- ARCPGA2023010005866_6378/Fondation ARC pour la Recherche sur le Cancer (ARC Foundation for Cancer Research)
- ERC StG TargetSWitch-101077864/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- ERC StG TargetSWitch-101077864/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

