A data assimilation framework for predicting the spatiotemporal response of high-grade gliomas to chemoradiation
- PMID: 40730976
- PMCID: PMC12308963
- DOI: 10.1186/s12885-025-14557-3
A data assimilation framework for predicting the spatiotemporal response of high-grade gliomas to chemoradiation
Abstract
Background: High-grade gliomas are highly invasive and respond variably to chemoradiation. Accurate, patient-specific predictions of tumor response could enhance treatment planning. We present a novel computational platform that assimilates MRI data to continually predict spatiotemporal tumor changes during chemoradiotherapy.
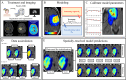
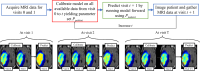
Methods: Tumor growth and response to chemoradiation was described using a two-species reaction-diffusion model of enhancing and non-enhancing regions of the tumor. Two evaluation scenarios were used to test the predictive accuracy of this model. In scenario 1, the model was calibrated on a patient-specific basis (n = 21) to weekly MRI data during the course of chemoradiotherapy. A data assimilation framework was used to update model parameters with each new imaging visit which were then used to update model predictions. In scenario 2, we evaluated the predictive accuracy of the model when fewer data points are available by calibrating the same model using only the first two imaging visits and then predicted tumor response at the remaining five weeks of treatment. We investigated three approaches to assign model parameters for scenario 2: (1) predictions using only parameters estimated by fitting the data obtained from an individual patient's first two imaging visits, (2) predictions made by averaging the patient-specific parameters with the cohort-derived parameters, and (3) predictions using only cohort-derived parameters.
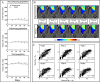
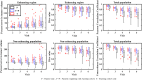
Results: Scenario 1 achieved a median [range] concordance correlation coefficient (CCC) between the predicted and measured total tumor cell counts of 0.91 [0.84, 0.95], and a median [range] percent error in tumor volume of -2.6% [-19.7, 8.0%], demonstrating strong agreement throughout the course of treatment. For scenario 2, the three approaches yielded CCCs of: (1) 0.65 [0.51, 0.88], (2) 0.74 [0.70, 0.91], (3) 0.76 [0.73, 0.92] with significant differences between the approach (1) that does not use the cohort parameters and the two approaches (2 and 3) that do.
Conclusions: The proposed data assimilation framework enhances the accuracy of tumor growth forecasts by integrating patient-specific and cohort-based data. These findings show a practical method for identifying more personalized treatment strategies in high-grade glioma patients.
Keywords: Computational oncology; Glioma; Imaging; Mathematical modeling; Personalized medicine.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All the patients involved in this study were enrolled on an institutional review board approved prospective study at M.D. Anderson Cancer Center (NCT# 04771806). All research methods adhered to the proper guidelines and regulations. Informed consent was collected from eligible patients that were enrolled into this trial. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–96. - PubMed
-
- Aum DJ, et al. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg Focus. 2014;37(6):E11. - PubMed
-
- Omuro A. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–50. - PubMed
-
- Ahmad SS, et al. Advances in radiotherapy. BMJ. 2012;345:e7765. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

