Artificial chromosome reorganization reveals high plasticity of the budding and fission yeast genomes
- PMID: 40731013
- PMCID: PMC12309097
- DOI: 10.1186/s13059-025-03689-1
Artificial chromosome reorganization reveals high plasticity of the budding and fission yeast genomes
Abstract
Background: The genome of a eukaryotic cell is usually organized on a set of chromosomes. Recently, karyotype engineering has been applied to various organisms, but whether and to what extent a naturally evolved genome can resist or tolerate massive artificial manipulations remains unexplored.
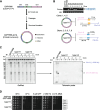
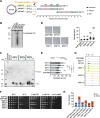
Results: Using unicellular yeast models of both Saccharomyces cerevisiae and Schizosaccharomyces pombe, we deliberately construct dozens of single-chromosome strains with different chromosome architectures. Three S. cerevisiae strains have the individual chromosomes fused into a single chromosome, but with the individual chromosomes in different orders. Eighteen S. cerevisiae strains have a single chromosome but with different centromeric sequences. Fifteen S. cerevisiae strains have a single chromosome with the centromere at different distances relative to the telomeres. Two S. pombe strains have a single, circular chromosome, and three strains have a single, linear chromosome with the centromere at different distances relative to the telomeres. All of these single-chromosome strains are viable, but the strains with an acrocentric or a telocentric chromosome have abnormal cell morphologies, and grow more slowly than those with a metacentric or sub-metacentric chromosome, and show increased genome instability with chromosome segregation abnormalities or genome diploidization.
Conclusion: The functional genomes of both the evolutionarily distant yeasts S. cerevisiae and S. pombe are highly tolerant of diversified genome organizations. The phenotypic abnormalities and increased genome instability of the acrocentric/telocentric single-chromosome yeasts suggest that yeasts with metacentric chromosomes have an evolutionary advantage.
Keywords: Chromosome engineering; Genome plasticity; Single-chromosome yeast; Telocentric chromosome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Ethical approval is not applicable for the study. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures





Similar articles
-
Plasticity of the mitotic spindle in response to karyotype variation.Curr Biol. 2024 Aug 5;34(15):3416-3428.e4. doi: 10.1016/j.cub.2024.06.058. Epub 2024 Jul 22. Curr Biol. 2024. PMID: 39043187 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Replication program of a single-chromosome budding yeast strain.Nucleic Acids Res. 2025 Aug 11;53(15):gkaf754. doi: 10.1093/nar/gkaf754. Nucleic Acids Res. 2025. PMID: 40808303 Free PMC article.
-
Centromeres in the thermotolerant yeast K. marxianus mediate attachment to a single microtubule.Chromosome Res. 2025 Jul 3;33(1):14. doi: 10.1007/s10577-025-09772-4. Chromosome Res. 2025. PMID: 40608157 Free PMC article.
-
Comparison of Telomere Structure in Eukaryotes.Arch Razi Inst. 2024 Dec 31;79(6):1365-1374. doi: 10.32592/ARI.2024.79.6.1365. eCollection 2024 Dec. Arch Razi Inst. 2024. PMID: 40606259 Free PMC article. Review.
References
-
- Uhler C, Shivashankar GV. Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat Rev Mol Cell Biol. 2017;18:717–27. - PubMed
-
- Talbert PB, Henikoff S. What makes a centromere? Exp Cell Res. 2020;389:111895. - PubMed
-
- Senaratne AP, Cortes-Silva N, Drinnenberg IA. Evolution of holocentric chromosomes: drivers, diversity, and deterrents. Semin Cell Dev Biol. 2022;127:90–9. - PubMed
-
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–80. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

