Endoplasmic reticulum-mitochondria coupling prompts ZBP1-mediated RPE cell PANoptosis in age-related macular degeneration
- PMID: 40731132
- PMCID: PMC12307978
- DOI: 10.1038/s42003-025-08565-z
Endoplasmic reticulum-mitochondria coupling prompts ZBP1-mediated RPE cell PANoptosis in age-related macular degeneration
Abstract
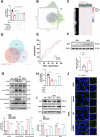
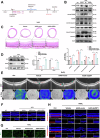
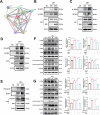
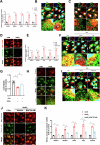
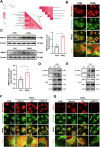
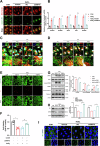
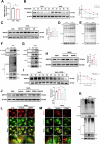
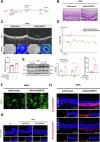
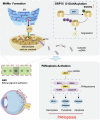
Age-related macular degeneration (AMD) is the leading cause of central vision impairment among the elderly. Geographic atrophy is a defining characteristic of AMD, but the detailed mechanism for massive loss of retinal pigment epithelium (RPE) cells is not fully understood. In this study, we found that Z-DNA binding protein 1 (ZBP1), a sensor for dsDNA, is able to induce RPE cell PANoptosis. Silencing ZBP1 efficiently alleviates RPE degeneration and AMD symptoms. Mechanistically, mitochondrial permeability transition pore (mPTP) opening stimulated by Ca2+ overload can trigger the releasing of mtDNA, which leads to ZBP1 activation and PANoptosis. Importantly, our findings reveal a significant role of aberrant formation of mitochondria-associated ER membranes (MAMs) in AMD. MAMs act as conduits for transferring Ca2+ from the ER to mitochondria through the VDAC1/GRP75/IP3R1 complex. Furthermore, our results indicate that GRP75 O-GlcNAcylation is involved in MAM formation. Genetic suppression of GRP75 attenuates PANoptosis and AMD progression. In summary, our study sheds light on the intricate organelle interplay underlying AMD and presents insights into potential avenues for AMD intervention.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
-
- Fleckenstein, M., Schmitz-Valckenberg, S. & Chakravarthy, U. Age-related macular degeneration: a review. JAMA331, 147–157 (2024). - PubMed
-
- Guymer, R. H. & Campbell, T. G. Age-related macular degeneration. Lancet401, 1459–1472 (2023). - PubMed
-
- Fleckenstein, M. et al. The progression of geographic atrophy secondary to age-related macular degeneration. Ophthalmology125, 369–390 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

