Mitochondrial complex I deficiency induces Alzheimer's disease-like signatures that are reversible by targeted therapy
- PMID: 40731203
- PMCID: PMC12307131
- DOI: 10.1002/alz.70519
Mitochondrial complex I deficiency induces Alzheimer's disease-like signatures that are reversible by targeted therapy
Abstract
Introduction: Mitochondrial dysfunction is implicated in Alzheimer's disease (AD), but whether it drives AD-associated changes is unclear. We assessed transcriptomic alterations in the brains of Ndufs4-/- mice, a model of mitochondrial complex I (mtCI) deficiency, and evaluated the therapeutic effects of the neuroprotective mtCI inhibitor CP2.
Methods: Cortico-hippocampal tissue from Ndufs4-/- and wild-type mice was subjected to transcriptomic analysis, followed by cross-species comparisons to human late-onset AD and familial AD mouse datasets.
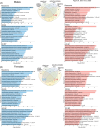
Results: Knockout of Ndufs4-mediated mtCI deficiency disrupted mitochondrial homeostasis, energy metabolism, and synaptic gene expression, recapitulating transcriptomic signatures of AD. CP2 treatment partially reversed these changes, with female Ndufs4-/- mice showing greater compensatory adaptations and treatment responses.
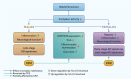
Discussion: Loss of mtCI activity alone is sufficient to induce AD-like molecular changes in the brain, independent of amyloid beta or phosphorylated tau. CP2-mediated rescue highlights the potential of targeting mitochondria as a therapeutic strategy for AD. Sex-specific responses suggest important considerations for personalized therapeutics.
Highlights: Activity of mitochondrial complex I (mtCI) affects broad mitochondrial and neuronal transcriptional networks. A reduction of mtCI activity is sufficient to induce transcriptomic changes reminiscent of those observed in late-onset Alsheimer's disease (AD) patients and familial mouse models of AD. Pharmacological targeting of mtCI mediates neuroprotective signaling. Male and female mice have differential responses to the loss of mtCI activity and to the mitochondria-targeted therapeutics. Mitochondria play a key role in AD development and treatment.
Keywords: Alzheimer's disease; Ndufs4 knockout mice; biological domains; mitochondrial complex I; mitochondria‐targeted therapeutics; mitophagy; sex‐specific differences; sex‐specific response; transcriptomic analysis; ubiquitin; weak complex I inhibitors.
© 2025 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
Dr. Eugenia Trushina and the Mayo Clinic hold four US patents on novel small‐molecule mitochondrion‐targeted compounds. Dr. Cory Funk is a cofounder of Fulcrum Neuroscience. Declarations of interest: None. Author disclosures are available in the supporting information.
Figures






References
-
- Swerdlow RH. The Alzheimer's disease mitochondrial cascade hypothesis: a current overview. J Alzheimers Dis. 2023;92:751‐768. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- RF1AG55549/NH/NIH HHS/United States
- Michael J. Fox Foundation for Parkinson's Research
- W81XWH-17-1-0248/Department of Defense Congressionally Directed Medical Research Programs
- R01 NS131322/NS/NINDS NIH HHS/United States
- RF1 AG062135/AG/NIA NIH HHS/United States
- R01NS112381/NH/NIH HHS/United States
- R56 AG062556/AG/NIA NIH HHS/United States
- Ted Nash Long Life Foundation
- R01NS110085/NH/NIH HHS/United States
- RO1AG062135/NH/NIH HHS/United States
- U54 AG065187/AG/NIA NIH HHS/United States
- RO1AG59093/NH/NIH HHS/United States
- R01NS131322/NH/NIH HHS/United States
- U54NS110435/NH/NIH HHS/United States
- R56AG062556/NH/NIH HHS/United States
- RF1NS085070/NH/NIH HHS/United States
- Emory-Sage-SGC-JAX U54AG065187/National Institute of Health Target Enablement to Accelerate Therapy Development for Alzheimer's Disease Consortia
- R01 NS110085/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

