Comprehensive analysis of the microbial consortium in the culture of flagellate Monocercomonoides exilis
- PMID: 40731366
- PMCID: PMC12308965
- DOI: 10.1186/s40793-025-00758-7
Comprehensive analysis of the microbial consortium in the culture of flagellate Monocercomonoides exilis
Abstract
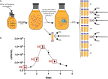
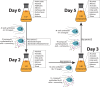
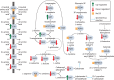
Monocercomonoides exilis is a model species of the amitochondrial eukaryotic group Oxymonadida, which makes it a suitable organism for studying the consequences of mitochondrial loss. Although M. exilis has an endobiotic lifestyle, it can be cultured in vitro in polyxenic conditions alongside an uncharacterized prokaryotic community, while attempts to create axenic cultures have not been successful. In this study, we used metagenomic sequencing, transcriptomics, and metabolomics to characterize the microbial consortium that supports the growth of M. exilis. We assembled genomes for 24 bacterial species and identified at least 30 species in total. M. exilis accounted for less than 1.5% of the DNA reads, while bacterial species dominated the sequence data and shifted in abundance over time. Our metabolic reconstruction and differential gene expression analyses show that the bacterial community relies on organic carbon oxidation, fermentation, and hydrogen production, but does not engage in methanogenesis. We observed rapid depletion of amino acids, nucleotides, glyceraldehyde, lactate, fatty acids, and alcohols in the medium, indicating a reliance on external nutrient recycling. The nitrogen cycle in this community is incomplete, with limited nitrogen fixation and no ammonia oxidation. Despite detailed metabolic profiling, we did not find any direct biochemical connections between M. exilis and the prokaryotes. Several bacterial species produce siderophores to assist themselves and others in the community in acquiring iron. However, M. exilis does not appear to benefit directly from siderophore-mediated iron transport and lacks known iron uptake pathways. This indicates that M. exilis may rely indirectly on the iron metabolism of other bacteria through phagocytosis. Additionally, some bacteria synthesize polyamines like spermidine and phosphatidylcholine, which M. exilis may need but cannot produce on its own. As the culture ages, M. exilis shows changes in gene expression consistent with starvation responses, including the upregulation of carbohydrate storage pathways and processes related to exocytosis. These findings provide new insights into microbial interactions within xenic cultures and emphasize the complex nature of maintaining amitochondriate eukaryotes in vitro.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interest: The authors declare no competing interests.
Figures



Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2020 Jan 9;1(1):CD011535. doi: 10.1002/14651858.CD011535.pub3. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Apr 19;4:CD011535. doi: 10.1002/14651858.CD011535.pub4. PMID: 31917873 Free PMC article. Updated.
References
-
- Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19:55–71. - PubMed
-
- Partida-Rodriguez O, Nieves-Ramirez M, Laforest-Lapointe I, Brown EM, Parfrey L, Valadez-Salazar A, et al. Exposure to parasitic protists and helminths changes the intestinal community structure of bacterial communities in a cohort of mother-child binomials from a semirural setting in Mexico. mSphere. 2021. 10.1128/mSphere.00083-21. - PMC - PubMed
Grants and funding
- 771592/H2020 European Research Council
- 771592/H2020 European Research Council
- 771592/H2020 European Research Council
- 771592/H2020 European Research Council
- 771592/H2020 European Research Council
- 771592/H2020 European Research Council
- 771592/H2020 European Research Council
- CZ.02.1.01/0.0/0.0/16_019/0000759/Centre for Research of Pathogenicity and Virulence of Parasites
- CZ.02.1.01/0.0/0.0/16_019/0000759/Centre for Research of Pathogenicity and Virulence of Parasites
- CZ.02.1.01/0.0/0.0/16_019/0000759/Centre for Research of Pathogenicity and Virulence of Parasites
- CZ.02.1.01/0.0/0.0/16_019/0000759/Centre for Research of Pathogenicity and Virulence of Parasites
- CZ.02.1.01/0.0/0.0/16_019/0000759/Centre for Research of Pathogenicity and Virulence of Parasites
- CZ.02.1.01/0.0/0.0/16_019/0000759/Centre for Research of Pathogenicity and Virulence of Parasites
- CZ.02.1.01/0.0/0.0/16_019/0000759/Centre for Research of Pathogenicity and Virulence of Parasites
LinkOut - more resources
Full Text Sources
Research Materials
