Spectral Precision: Recent Advances in Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry for Pathogen Detection and Resistance Profiling
- PMID: 40731982
- PMCID: PMC12299549
- DOI: 10.3390/microorganisms13071473
Spectral Precision: Recent Advances in Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry for Pathogen Detection and Resistance Profiling
Abstract
With the global rise in antimicrobial resistance (AMR), rapid and reliable microbial diagnostics have become more critical than ever. Traditional culture-based and molecular diagnostic techniques often fall short in terms of speed, cost-efficiency, or scalability, particularly in resource-limited settings. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has emerged as a transformative tool in clinical microbiology. Its unparalleled speed and accuracy in microbial identification, along with expanding applications in AMR profiling, make it a leading candidate for next-generation diagnostic workflows. This review aims to provide a comprehensive update on recent advances in MALDI-TOF MS, focusing on its technological evolution, clinical applications, and future potential in microbial diagnostics and resistance detection. We conducted a critical synthesis of peer-reviewed literature published over the last decade, with emphasis on innovations in sample preparation, instrumentation, data interpretation, and clinical integration. Key developments in AMR detection, including growth-based assays, resistance biomarker profiling, and machine learning-driven spectral analysis, are discussed. MALDI-TOF MS is increasingly deployed not only in clinical laboratories but also in environmental surveillance, food safety, and military biodefense. Despite challenges such as database variability and limited access in low-income regions, it remains a cornerstone of modern microbial diagnostics and holds promise for future integration into global AMR surveillance systems.
Keywords: MALDI–TOF MS; antibiotic resistance; machine learning; microbial diagnostics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

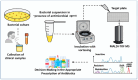

References
-
- Wat J.K.-H., Xu M., Nan L., Lin H., To K.K.-W., Shum H.C., Hassan S.U. Rapid antimicrobial susceptibility tests performed by self-diluting microfluidic chips for drug resistance studies and point-of-care diagnostics. Microsyst. Nanoeng. 2025;11:110. doi: 10.1038/s41378-025-00938-y. - DOI - PMC - PubMed
-
- Mwaturura T., Olaru I.D., Chimhini G., Bwakura-Dangarembizi M., Mangiza M., Chimhuya S., Sado B., Katunga J., Tarupiwa A., Juru A. Rapid bacterial identification and resistance detection using a low complexity molecular diagnostic platform in Zimbabwe. PLOS Glob. Public Health. 2025;5:e0004343. doi: 10.1371/journal.pgph.0004343. - DOI - PMC - PubMed
-
- Church D.L., Cerutti L., Gürtler A., Griener T., Zelazny A., Emler S. Performance and application of 16S rRNA gene cycle sequencing for routine identification of bacteria in the clinical microbiology laboratory. Clin. Microbiol. Rev. 2020;33:e00053-19. doi: 10.1128/CMR.00053-19. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

