Peculiarities of Diagnostic Reliability-Nested PCR Versus SAT in the Identification of Helicobacter pylori
- PMID: 40732007
- PMCID: PMC12301005
- DOI: 10.3390/microorganisms13071498
Peculiarities of Diagnostic Reliability-Nested PCR Versus SAT in the Identification of Helicobacter pylori
Abstract
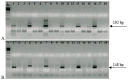
H. pylori detection via the stool antigen test (SAT) requires 100 times more cells than nested PCR (NPCR) for a 454 bp amplicon, but is significantly more sensitive in identifying positive stool samples. To understand this contradiction, we developed an NPCR assay to amplify a shorter 148 bp segment of the 16S rRNA gene. The assay was extremely sensitive and reliable when adhering to particular rules commonly used in forensic laboratories. The SAT and NPCR for long and short amplicons were compared using stool samples from 208 gastroenterological patients, of which 27.9% were identified as positive according to the SAT and only 6.25% according to the 454 bp NPCR amplicon, but 51.0% in the short 148 bp NPCR. Among 100 asymptomatic volunteers, the prevalence was 35% in the SAT assay and 22% in the long NPCR, but as much as 66.6% of positives were determined in the short 148 bp NPCR. The specificity of the PCR product was determined via DNA sequencing, which confirmed H. pylori's origin in all NPCR-positive samples. Apparently, the stool contains mostly short fragments of H. pylori DNA, and the most plausible explanation for the SAT/NPCR paradox is the degradation of H. pylori DNA in the digestive system.
Keywords: H. pylori; diagnostic reliability; nested PCR; stool antigen test.
Conflict of interest statement
Author Jana Krčmáriková was employed by the company Synlab Slovakia s. r. o. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Non-invasive diagnostic tests for Helicobacter pylori infection.Cochrane Database Syst Rev. 2018 Mar 15;3(3):CD012080. doi: 10.1002/14651858.CD012080.pub2. Cochrane Database Syst Rev. 2018. PMID: 29543326 Free PMC article.
-
DNA diagnostics for reliable and universal identification of Helicobacter pylori.World J Gastroenterol. 2021 Nov 7;27(41):7100-7112. doi: 10.3748/wjg.v27.i41.7100. World J Gastroenterol. 2021. PMID: 34887630 Free PMC article. Review.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
-
Optimum duration of regimens for Helicobacter pylori eradication.Cochrane Database Syst Rev. 2013 Dec 11;2013(12):CD008337. doi: 10.1002/14651858.CD008337.pub2. Cochrane Database Syst Rev. 2013. PMID: 24338763 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

