Gut Microbiome Modulation and Health Benefits of a Novel Fucoidan Extract from Saccharina latissima: A Double-Blind, Placebo-Controlled Trial
- PMID: 40732054
- PMCID: PMC12298129
- DOI: 10.3390/microorganisms13071545
Gut Microbiome Modulation and Health Benefits of a Novel Fucoidan Extract from Saccharina latissima: A Double-Blind, Placebo-Controlled Trial
Abstract
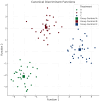
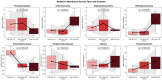
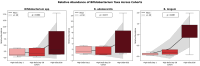
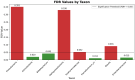
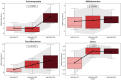
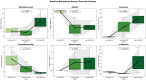
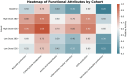
This randomized, double-blind, placebo-controlled, three-arm clinical trial evaluated the effects of a proprietary bioactive fucoidan-rich extract derived from Saccharina latissima (SLE-F) on gut microbial composition and function in healthy adults. The objective of the study was to assess the potential of SLE-F to beneficially modulate the gut microbiome, with this paper specifically reporting on microbial diversity, taxonomic shifts, and functional pathway outcomes. Ninety-one participants received either a low dose (125 mg), high dose (500 mg), or placebo twice daily for four weeks. The primary endpoint was the microbiome composition assessed via 16S rRNA sequencing (V3-V4 region), with secondary outcomes including surveys, adverse event monitoring, and clinical evaluations. High-dose supplementation resulted in dose-dependent improvements in the microbial diversity; increased abundance of beneficial taxa, including Bifidobacterium, Faecalibacterium, and Lachnospiraceae; and reductions in inflammation-associated taxa, such as Enterobacteriaceae and Pseudomonadota. A functional pathway analysis showed enhancement in short-chain fatty acid biosynthesis and carbohydrate metabolism. The low-dose group showed modest benefits, primarily increasing Bifidobacterium, with limited functional changes. In vitro colonic simulations further demonstrated a dose-dependent increase in short-chain fatty acids and postbiotic metabolite production following SLE-F exposure. SLE-F was well tolerated, with only mild, nonspecific adverse events reported. These findings support the potential of SLE-F as a safe and effective microbiome-modulating agent, warranting further study of the long-term use and synergy with dietary interventions.
Keywords: SCFA; Saccharina latissima; clinical trial; fucoidan; intestinal transit; microbiome.
Conflict of interest statement
Author Charles Bavington, Raminta Kazlauskaite, and Neil Waslidge are employed by the company Oceanium, Ltd. The authors declare that this study received funding from Oceanium, Ltd. The funder was not involved in the study design, collection, analysis, interpretation of data; the writing of this article; or the decision to submit it for publication. All other authors claim no conflict of interest.
Figures


Similar articles
-
Characterizing Gene-Level Adaptations in the Gut Microbiome During Viral Infections: The Role of a Fucoidan-Rich Extract.Genes (Basel). 2025 Jun 26;16(7):740. doi: 10.3390/genes16070740. Genes (Basel). 2025. PMID: 40725397 Free PMC article.
-
Effects of supplementation of live and heat-treated Bifidobacterium animalis subspecies lactis CECT 8145 on glycemic and insulinemic response, fecal microbiota, systemic biomarkers of inflammation, and white blood cell gene expression of adult dogs.J Anim Sci. 2024 Jan 3;102:skae291. doi: 10.1093/jas/skae291. J Anim Sci. 2024. PMID: 39320367
-
Gut microbiome-based interventions for the management of obesity in children and adolescents aged up to 19 years.Cochrane Database Syst Rev. 2025 Jul 10;7(7):CD015875. doi: 10.1002/14651858.CD015875. Cochrane Database Syst Rev. 2025. PMID: 40637175 Review.
-
Corticosteroids for the treatment of Duchenne muscular dystrophy.Cochrane Database Syst Rev. 2016 May 5;2016(5):CD003725. doi: 10.1002/14651858.CD003725.pub4. Cochrane Database Syst Rev. 2016. PMID: 27149418 Free PMC article.
-
Oral herbal therapies for treating osteoarthritis.Cochrane Database Syst Rev. 2014 May 22;2014(5):CD002947. doi: 10.1002/14651858.CD002947.pub2. Cochrane Database Syst Rev. 2014. PMID: 24848732 Free PMC article.
References
-
- Zhou Y., Xu Z., Zhang H., Liu T., Zhou J., Bi Y., Han Y., Tan Y., Yuan J., Yang R. When the microbiome meets One Health principle: Leading to the Holy Grail of biology and contributing to overall well-being and social sustainability. iMetaOmics. 2024;1:e30. doi: 10.1002/imo2.30. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

