The Microbiological Characteristics and Genomic Surveillance of Carbapenem-Resistant Klebsiella pneumoniae Isolated from Clinical Samples
- PMID: 40732086
- PMCID: PMC12299758
- DOI: 10.3390/microorganisms13071577
The Microbiological Characteristics and Genomic Surveillance of Carbapenem-Resistant Klebsiella pneumoniae Isolated from Clinical Samples
Abstract
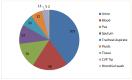
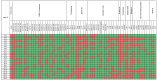
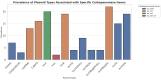
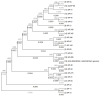
Klebsiella pneumoniae is a major public health concern due to its role in Gram-negative bacteremia, which leads to high mortality and increased healthcare costs. This study characterizes phenotypic and genomic features of K. pneumoniae isolates from clinical samples in Karachi, Pakistan. Among 507 isolates, 213 (42%) were carbapenem-resistant based on disk diffusion and MIC testing. Urine (29.7%) and blood (28.3%) were the most common sources, with infections predominantly affecting males (64.7%) and individuals aged 50-70 years. Colistin was the only antibiotic showing consistent activity against these isolates. The whole-genome sequencing of 24 carbapenem-resistant K. pneumoniae (CR-KP) isolates revealed blaNDM-5 (45.8%) as the dominant carbapenemase gene, followed by blaNDM-1 (12.5%) and blaOXA-232 (54.2%). Other detected blaOXA variants included blaOXA-1, blaOXA-4, blaOXA-10, and blaOXA-18. The predominant beta-lactamase gene was blaCTX-M-15 (91.6%), followed by blaCTX-M-163, blaCTX-M-186, and blaCTX-M-194. Sequence types ST147, ST231, ST29, and ST11 were associated with resistance. Plasmid profiling revealed IncR (61.5%), IncL (15.4%), and IncC (7.7%) as common plasmid types. Importantly, resistance was driven not only by acquired genes but also by chromosomal mutations. Porin mutations in OmpK36 and OmpK37 (e.g., P170M, I128M, N230G, A217S) reduced drug influx, while acrR and ramR mutations (e.g., P161R, G164A, P157*) led to efflux pump overexpression, enhancing resistance to fluoroquinolones and tigecycline. These findings highlight a complex resistance landscape driven by diverse carbapenemases and ESBLs, underlining the urgent need for robust antimicrobial stewardship and surveillance strategies.
Keywords: antimicrobial resistance; carbapenem resistant K. pneumoniae; molecular epidemiology; whole genome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Sisay A., Kumie G., Gashaw Y., Nigatie M., Gebray H.M., Reta M.A. Prevalence of genes encoding carbapenem-resistance in Klebsiella pneumoniae recovered from clinical samples in Africa: Systematic review and meta-analysis. BMC Infect. Dis. 2025;25:556. doi: 10.1186/s12879-025-10959-7. - DOI - PMC - PubMed
-
- Chaudhari V.D.C., Kumar M., Das N.G.N. AmpC and extended-spectrum beta lactamase producing isolates of E. coli, Klebsiella spp. and P. mirabilis in a tertiary care center and their sensitivity to other antibiotics. Int. J. Curr. Microbiol. Appl. Sci. 2017;6:868–877. doi: 10.20546/ijcmas.2017.603.102. - DOI
-
- Casale R., Boattini M., Comini S., Bastos P., Corcione S., De Rosa F.G., Bianco G., Costa C. Clinical and microbiological features of positive blood culture episodes caused by non-fermenting gram-negative bacilli other than Pseudomonas and Acinetobacter species (2020–2023) Infection. 2024;53:183–196. doi: 10.1007/s15010-024-02342-6. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

