Identification and Pathogenicity Analysis of Huaxiibacter chinensis Qf-1 in Mink (Neogale vison)
- PMID: 40732113
- PMCID: PMC12300476
- DOI: 10.3390/microorganisms13071604
Identification and Pathogenicity Analysis of Huaxiibacter chinensis Qf-1 in Mink (Neogale vison)
Abstract
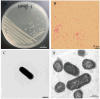
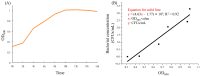
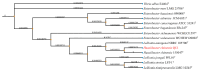
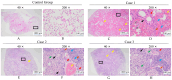
Mink (Neogale vison) is a commercially farmed animal of global importance. However, disease outbreaks during farming not only cause significant economic losses but also substantially increase the risk of zoonotic infections. The identification and characterization of pathogenic bacteria remain a major bottleneck restricting the development of healthy and sustainable mink farming. In this study, an LB medium was used to isolate a pale-white, rod-shaped, Gram-negative bacterial strain, Qf-1, from minks with pneumonia. Based on morphological characteristics, biochemical properties, 16S rRNA gene sequencing, and average nucleotide identity (ANI) analysis, strain Qf-1 was identified as Huaxiibacter chinensis Qf-1. Under laboratory conditions, H. chinensis Qf-1 induced typical pneumonia symptoms in Kunming mice. Furthermore, whole-genome sequencing of H. chinensis Qf-1 revealed its genome to be 4.77 Mb and to contain a single chromosome and one plasmid. The main virulence genes of H. chinensis Qf-1 were primarily associated with flgB, flgC, flgG, aceA, hemL, tssC1, csgD, hofB, ppdD, hcpA, and vgrGA, functioning in motility, biofilm formation, colonization ability, and secretion systems. Our findings contribute to a better understanding of their pathogenic mechanisms, thereby laying a theoretical foundation for further investigation into the complex interactions between gut microbiota and the host.
Keywords: Huaxiibacter chinensis Qf-1; Neogale vison; culturomics; genome; pathogenicity.
Conflict of interest statement
Author Shuli Liu was employed by the company Zhonghuan Shengda Environmental Technology Group (Qingyun) Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
-
- Yu Y., Hu B., Fan H., Zhang H., Lian S., Li H., Li S., Yan X., Wang S., Bai X. Molecular Epidemiology of Extraintestinal Pathogenic Escherichia coli Causing Hemorrhagic Pneumonia in Mink in Northern China. Front. Cell. Infect. Microbiol. 2021;11:781068. doi: 10.3389/fcimb.2021.781068. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

