Multi-Omics Integration Reveals the Impact of Gastrointestinal Microbiota on Feed Efficiency in Tan Sheep
- PMID: 40732118
- PMCID: PMC12298153
- DOI: 10.3390/microorganisms13071608
Multi-Omics Integration Reveals the Impact of Gastrointestinal Microbiota on Feed Efficiency in Tan Sheep
Abstract
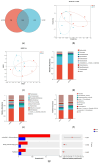
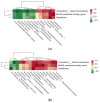
The rumen and intestinal microbiota play a pivotal role in the digestion and absorption processes of ruminants. Elucidating the mechanisms by which gastrointestinal microbiota influence the feed conversion ratio (FCR) in ruminants is significantly important for enhancing feed utilization efficiency in these animals. In this study, RT-qPCR, 16S rRNA sequencing, and metabolomic techniques were systematically employed to compare the microbial community structures in the rumen, cecum, and rectum, as well as the differences in rumen metabolites between high- and low-FCR Tan sheep. The results showed that, compared to the HFCR group of Tan sheep, the LFCR group exhibited a significant reduction in unclassified_f__Selenomonadaceae, Blvii28_wastewater-sludge_group, and Papillibacter in the rumen; a significant increase in Lachnospiraceae_AC2044_group and Sanguibacteroides; a significant reduction in unclassified_f__Peptostreptococcaceae, Clostridium_sensu_stricto_1, and Parasutterella in the cecum; a significant increase in norank_f__Bacteroidales_UCG-001; and a significant reduction in norank_f__Muribaculaceae, Blautia, and Turicibacter in the rectum. There is a significant positive correlation between Parasutterella in the cecum and three microorganisms, including unclassified_f__Selenomonadaceae, in the rumen. Additionally, Blvii28_wastewater-sludge_group was positively correlated with Lactobacillus. Furthermore, unclassified_f__Selenomonadaceae in the rumen was positively correlated with Turicibacter, unclassified_f__Peptostreptococcaceae, and Breznakia in the rectum. Blvii28_wastewater-sludge_group also showed positive correlations with Blautia, norank_f__Muribaculaceae, and Clostridium_sensu_stricto_1, while Papillibacter was positively correlated with Faecalitalea. The metabolomic results indicated that, compared to the HFCR group, 261 differential metabolites, including Phenylacetylglutamine and Populin, in the rumen of Tan sheep in the LFCR group were significantly downregulated, whereas 36 differential metabolites, including Glycyl-L-tyrosine, were significantly upregulated. Furthermore, the rumen microbe unclassified_f__Selenomonadaceae exhibited positive correlations with significantly differential metabolites such as L-tryptophan, Etiocholanolone glucuronide, N-acetyl-O-demethylpuromycin, and 6-deoxyerythronolide B. Blvii28_wastewater-sludge_group and Papillibacter also exhibited positive correlations with Icilin. High and low FCRs in the rumen of Tan sheep were investigated, especially in relation to unclassified_f__Selenomonadaceae, Blvii28_wastewater-sludge_group, and Papillibacter. Correlations can be seen with microorganisms such as Parasutatella and Lactobacillus in the cecum; Turicibacter, norank_f__Bacteroideales_UCG-001, and Blautia in the rectum; and metabolites such as L-tryptophan, Etiocholanolone glucuronide, and N-acetyl-O-demethylpuromycin. This reveals the role of microorganisms in the digestion and absorption of Tan sheep feed, thus providing a preliminary basis for further research on the microbial regulation of ruminant animal feed utilization and a theoretical basis for improving Tan sheep feed utilization efficiency.
Keywords: 16S rRNA; cecum; metabolomics; rectum; rumen.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Effects of starter diets with different neutral detergent fiber levels on gastrointestinal tract development, microbial profiles, nutrient digestibility, and growth performance in Xinong Saanen dairy goat kids.J Anim Sci. 2025 Jan 4;103:skaf226. doi: 10.1093/jas/skaf226. J Anim Sci. 2025. PMID: 40650561
-
Effects of extruded corn distillers dried grains with solubles on growth performance, nutrient digestibility, gut health, and microbiota diversity in weaned piglets.J Anim Sci. 2025 Jan 4;103:skaf120. doi: 10.1093/jas/skaf120. J Anim Sci. 2025. PMID: 40244394
-
Integrative multi-omics and bioinformatics analysis of the effects of BaiRui YuPingFeng Powder on intestinal health in broilers.Front Vet Sci. 2025 Jun 18;12:1606531. doi: 10.3389/fvets.2025.1606531. eCollection 2025. Front Vet Sci. 2025. PMID: 40607343 Free PMC article.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
References
-
- Zhao X., Zuo S., Guo Y., Zhang C., Wang Y., Peng S., Liu M., Wang B., Zhang H., Luo H. Carcass Meat Quality, Volatile Compound Profile, and Gene Expression in Tan Sheep under Different Feeding Regimes. Food Biosci. 2023;56:103213. doi: 10.1016/j.fbio.2023.103213. - DOI
-
- Lu M.-L., Pan L., Zheng C., Mao R.-Y., Yuan G.-H., Shi C.-Y., Pu Z.-H., Su H.-X., Diao Q.-Y., Rehemujiang H., et al. Effects of Compound Microecological Preparation Supplementation on Production Performance and Nutrient Apparent Digestibility in Hu Sheep from the Rumen Perspective. Microorganisms. 2025;13:999. doi: 10.3390/microorganisms13050999. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

