L-Arabinose Alters the E. coli Transcriptome to Favor Biofilm Growth and Enhances Survival During Fluoroquinolone Stress
- PMID: 40732174
- PMCID: PMC12299780
- DOI: 10.3390/microorganisms13071665
L-Arabinose Alters the E. coli Transcriptome to Favor Biofilm Growth and Enhances Survival During Fluoroquinolone Stress
Abstract
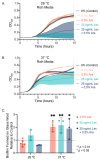
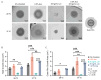
Environmental conditions, including nutrient composition and temperature, influence biofilm formation and antibiotic resistance in Escherichia coli. Understanding how specific metabolites modulate these processes is critical for improving antimicrobial strategies. Here, we investigated the growth and composition of Escherichia coli in both planktonic and biofilm states in the presence of L-arabinose, with and without exposure to the fluoroquinolone antibiotic levofloxacin, at two temperatures: 28 and 37 °C. At both temperatures, L-arabinose increased the growth rate of planktonic E. coli but resulted in reduced total growth; concurrently, it enhanced biofilm growth at 37 °C. L-arabinose reduced the efficacy of levofloxacin and promoted growth in sub-minimum inhibitory concentrations (25 ng/mL). Transcriptomic analyses provided insight into the molecular basis of arabinose-mediated reduced susceptibility of E. coli to levofloxacin. We found that L-arabinose had a temperature- and state-dependent impact on the transcriptome. Using gene ontology overrepresentation analyses, we found that L-arabinose modulated the expression of many critical antibiotic resistance genes, including efflux pumps (ydeA, mdtH, mdtM), transporters (proVWX), and biofilm-related genes for external structures like pili (fimA) and curli (csgA, csgB). This study demonstrates a previously uncharacterized role for L-arabinose in modulating antibiotic resistance and biofilm-associated gene expression in E. coli and provides a foundation for additional exploration of sugar-mediated antibiotic sensitivity in bacterial biofilms.
Keywords: E. coli; biofilm; fluoroquinolone; transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Non-disruptive matrix turnover is a conserved feature of biofilm aggregate growth in paradigm pathogenic species.mBio. 2025 Mar 12;16(3):e0393524. doi: 10.1128/mbio.03935-24. Epub 2025 Feb 21. mBio. 2025. PMID: 39982068 Free PMC article.
-
Cefmetazole, flomoxef, and meropenem are effective against planktonic cells but not biofilms of extended-spectrum β-lactamase-producing Escherichia coli.BMC Microbiol. 2025 Jul 2;25(1):387. doi: 10.1186/s12866-025-04088-z. BMC Microbiol. 2025. PMID: 40604378 Free PMC article.
-
Multispecies biofilm architecture determines bacterial exposure to phages.PLoS Biol. 2022 Dec 22;20(12):e3001913. doi: 10.1371/journal.pbio.3001913. eCollection 2022 Dec. PLoS Biol. 2022. PMID: 36548227 Free PMC article.
-
Perioperative antibiotics for prevention of acute endophthalmitis after cataract surgery.Cochrane Database Syst Rev. 2017 Feb 13;2(2):CD006364. doi: 10.1002/14651858.CD006364.pub3. Cochrane Database Syst Rev. 2017. PMID: 28192644 Free PMC article.
-
Amnioinfusion for chorioamnionitis.Cochrane Database Syst Rev. 2016 Aug 24;2016(8):CD011622. doi: 10.1002/14651858.CD011622.pub2. Cochrane Database Syst Rev. 2016. PMID: 27556818 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

