High-Resolution Core Gene-Associated Multiple Nucleotide Polymorphism (cgMNP) Markers for Strain Identification in the Wine Cap Mushroom Stropharia rugosoannulata
- PMID: 40732194
- PMCID: PMC12298363
- DOI: 10.3390/microorganisms13071685
High-Resolution Core Gene-Associated Multiple Nucleotide Polymorphism (cgMNP) Markers for Strain Identification in the Wine Cap Mushroom Stropharia rugosoannulata
Abstract
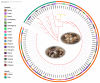
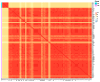
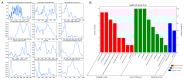
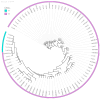
Stropharia rugosoannulata, an ecologically valuable and economically important edible mushroom, faces challenges in strain-level identification and breeding due to limited genomic resources and the lack of high-resolution molecular markers. In this study, we generated high-quality genomic data for 105 S. rugosoannulata strains and identified over 2.7 million SNPs, unveiling substantial genetic diversity within the species. Using core gene-associated multiple nucleotide polymorphism (cgMNP) markers, we developed an efficient and transferable framework for strain discrimination. The analysis revealed pronounced genetic differentiation among cultivars, clustering them into two distinct phylogenetic groups. Nucleotide diversity (π) across 83 core genes varied significantly, highlighting both highly conserved loci under purifying selection and highly variable loci potentially associated with adaptive evolution. Phylogenetic analysis of the most variable gene, Phosphatidate cytidylyltransferase mitochondrial, identified 865 SNPs, enabling precise differentiation of all 85 cultivars. Our findings underscore the utility of cgMNP markers in addressing challenges posed by horizontal gene transfer and phylogenetic noise, demonstrating their robustness in cross-species applications. By providing insights into genetic diversity, evolutionary dynamics, and marker utility, this study establishes a foundation for advancing breeding programs, conservation strategies, and functional genomics in S. rugosoannulata. Furthermore, the adaptability of cgMNP markers offers a universal tool for high-resolution strain identification across diverse fungal taxa, contributing to broader fungal phylogenomics and applied mycology.
Keywords: core gene-associated MNP markers; phylogenetic tree; phylogenomics; strain identification.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
TEA5K: a high-resolution and liquid-phase multiple-SNP array for molecular breeding in tea plant.J Nanobiotechnology. 2025 Jul 2;23(1):481. doi: 10.1186/s12951-025-03533-5. J Nanobiotechnology. 2025. PMID: 40605004 Free PMC article.
-
The rise of Stropharia rugosoannulata industry in China: current state and prospects.Appl Microbiol Biotechnol. 2025 Aug 20;109(1):188. doi: 10.1007/s00253-025-13576-1. Appl Microbiol Biotechnol. 2025. PMID: 40830602 Free PMC article. Review.
-
Candidate gene identification and marker development for seed coat peeling rate in peanut (Arachis Hypogaea L.).BMC Plant Biol. 2025 Jul 25;25(1):959. doi: 10.1186/s12870-025-07007-6. BMC Plant Biol. 2025. PMID: 40713477 Free PMC article.
-
Identification of stable reference genes for qRT-PCR in Stropharia rugosoannulata using mRNA-sequencing data.PLoS One. 2025 May 7;20(5):e0323272. doi: 10.1371/journal.pone.0323272. eCollection 2025. PLoS One. 2025. PMID: 40334243 Free PMC article.
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
References
-
- Yan Q.-X., Huang M.-X., Sun P., Cheng S., Zhang Q., Dai H. Steroids, Fatty Acids and Ceramide from the Mushroom Stropharia rugosoannulata Farlow apud Murrill. Biochem. Syst. Ecol. 2020;88:103963. doi: 10.1016/j.bse.2019.103963. - DOI
-
- Szudyga K. The Biology and Cultivation of Edible Mushrooms. Elsevier; Amsterdam, The Netherlands: 1978. Stropharia Rugoso-Annulata; pp. 559–571.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

