Synthesis and Biological Evaluation of Herceptin-Conjugated Liposomes Loaded with Lipocalin-2 siRNA for the Treatment of Inflammatory Breast Cancer
- PMID: 40732340
- PMCID: PMC12299312
- DOI: 10.3390/ph18071053
Synthesis and Biological Evaluation of Herceptin-Conjugated Liposomes Loaded with Lipocalin-2 siRNA for the Treatment of Inflammatory Breast Cancer
Abstract
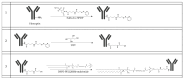
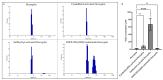
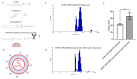
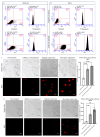
Background: Inflammatory breast cancer (IBC) is a rare and aggressive subtype of breast cancer that accounts for 1-5% of BC patients and regularly affects women under 40 years of age. Approximately 50% of IBC cases are HER2+ and can be treated with the monoclonal antibody-based therapy Herceptin (trastuzumab). However, resistance to Herceptin develops within a year, and effective second-line targeted therapies are currently unavailable for IBC patients. Lipocalin-2 (LCN2) is a promising therapeutic target for IBC due to its role in promoting tumor invasiveness, angiogenesis, and the inflammatory tumor microenvironment characteristic of IBC. Objective: We developed Herceptin-conjugated liposomes loaded with LCN2-targeted small-interference RNA (siRNA) for HER2+ IBCs. Methods: We synthesized DSPE-PEG(2000)-maleimide-Herceptin in a three-step process and formulated the liposomes together with DOPC, PEG(2000)-PE, cholesterol, and siRNA. Results: Dynamic light scattering confirmed the liposome size distribution, which was 66.7 nm for the Herceptin-conjugated liposome versus 43.0 nm in a non-functionalized liposome. Here, we report efficient internalization of this formulation into HER2+ IBC cells, reducing LCN2 levels by 30% and disrupting tumor emboli formation. RNA sequencing revealed 139 genes that were differentially expressed upon LCN2 knockdown, with 25 canonical pathways identified through Ingenuity Pathway Analysis. Conclusions: These findings suggest that LCN2-targeted siRNA within Herceptin-targeted liposomes represents a promising therapeutic strategy for IBC.
Keywords: inflammatory breast cancer; lipocalin-2; liposomes; siRNA; tumor emboli.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Chippa V., Barazi H. StatPearls[Internet] StatPearls Publishing; Treasure Island, FL, USA: 2023. [(accessed on 31 January 2025)]. Inflammatory Breast Cancer. Available online: https://www.ncbi.nlm.nih.gov/books/NBK564324/ - PubMed
-
- De Schepper M., Nguyen H.-L., Richard F., Rosias L., Lerebours F., Vion R., Clatot F., Berghian A., Maetens M., Leduc S., et al. Treatment Response, Tumor Infiltrating Lymphocytes and Clinical Outcomes in Inflammatory Breast Cancer–Treated with Neoadjuvant Systemic Therapy. Cancer Res. Commun. 2024;4:186–199. doi: 10.1158/2767-9764.CRC-23-0285. - DOI - PMC - PubMed
-
- National Cancer Institute Cancer Stat Facts: Female Breast Cancer. [(accessed on 28 March 2025)]; Available online: https://seer.cancer.gov/statfacts/html/breast.html.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

