Electroporation- and Liposome-Mediated Co-Transfection of Single and Multiple Plasmids
- PMID: 40733113
- PMCID: PMC12297979
- DOI: 10.3390/pharmaceutics17070905
Electroporation- and Liposome-Mediated Co-Transfection of Single and Multiple Plasmids
Abstract
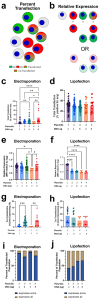
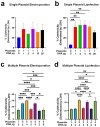
Background/Objectives: Co-transfection of multiple DNAs is important to many research and therapeutic applications. While the optimization of single plasmid transfection is common, multiple plasmid co-transfection analyses are limited. Here we provide empirical data regarding multiple plasmid co-transfection while altering the number of species of plasmids transfected (up to four different plasmids) and the amount of plasmids/cell using the two most common non-viral techniques, electroporation and lipofection. Methods: A549 human lung epithelial cells were transfected using lipofectamine 2000 or electroporation with combinations of plasmids, each expressing one of four different fluorescent proteins from the CAGG promoter. Twenty-four hours later, cells were analyzed by spectral flow cytometry to determine the number of cells expressing each fluorescent protein and the amount of fluorescent signal of each protein in a cell. Results and Conclusions: For electroporation, while the fraction of cells expressing plasmids increased with increasing amounts of DNA, increasing the number of plasmid species did not alter the fraction of expressing cells and had no effect on levels of expression in individual cells. By contrast, for lipofection, the fraction of cells expressing plasmids was not affected by the amount of DNA added but both the fraction of cells expressing and the level of protein produced in these cells decreased for each plasmid species as the number of delivered species increased. For both lipofection and electroporation after single plasmid transfection, the expressing cells had greater numbers of plasmid copies/cell than non-expressing cells. Multiple plasmid lipofection resulted in more plasmid copies/cell in co-expressing than non-expressing cells. Multiple plasmid electroporation was the inverse of this with fewer plasmid copies/cell in co-expressing than non-expressing cells.
Keywords: electroporation; gene delivery; gene electrotransfer; lipofection; multiple plasmids; plasmids; transfection.
Conflict of interest statement
David Dean serves as a member of the Scientific Advisory Board of Seawolf Therapeutics and has equity interest in the company. Uday Baliga, Anthony Gurunian, Aitor Nogales, and Luis Martinez-Sobrido have no conflicting interests to declare.
Figures


Similar articles
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Interventions for central serous chorioretinopathy: a network meta-analysis.Cochrane Database Syst Rev. 2025 Jun 16;6(6):CD011841. doi: 10.1002/14651858.CD011841.pub3. Cochrane Database Syst Rev. 2025. PMID: 40522203
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Audit and feedback: effects on professional practice.Cochrane Database Syst Rev. 2025 Mar 25;3(3):CD000259. doi: 10.1002/14651858.CD000259.pub4. Cochrane Database Syst Rev. 2025. PMID: 40130784
References
-
- Cardarelli F., Digiacomo L., Marchini C., Amici A., Salomone F., Fiume G., Rossetta A., Gratton E., Pozzi D., Caracciolo G. The intracellular trafficking mechanism of Lipofectamine-based transfection reagents and its implication for gene delivery. Sci. Rep. 2016;6:25879. doi: 10.1038/srep25879. - DOI - PMC - PubMed
-
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M., et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

