Genome-Wide Analysis of GmMYB S20 Transcription Factors Reveals Their Critical Role in Soybean Nodulation
- PMID: 40733477
- PMCID: PMC12298257
- DOI: 10.3390/plants14142240
Genome-Wide Analysis of GmMYB S20 Transcription Factors Reveals Their Critical Role in Soybean Nodulation
Abstract
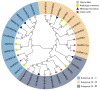
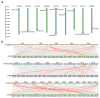
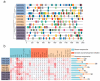
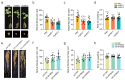
Soybean relies on symbiotic nitrogen fixation (SNF) to support sustainable agriculture. In this study, we conducted a comprehensive analysis of the GmMYB transcription factor subfamily 20, with a focus on GmMYB62a and GmMYB62b. Phylogenetic and structural analyses revealed that these genes are evolutionarily conserved among legumes and possess distinct domain architectures. Expression profiling and GUS staining showed that GmMYB62a and GmMYB62b are constitutively expressed in nodules. Functional analyses revealed that loss of GmMYB62s function significantly reduced nodule density, while overexpression promoted nodulation. Transcriptomic analysis (RNA-seq) further demonstrated that GmMYB62s regulate key pathways, including hormone signaling, immune responses, and cell wall metabolism, thereby coordinating symbiotic interactions. Collectively, our findings identify GmMYB62a and GmMYB62b as critical molecular regulators of nodulation in soybean, providing promising targets for improving symbiotic nitrogen fixation efficiency in legume crops.
Keywords: GmMYB62a and GmMYB62b; nodulation; soybean; transcription factor.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Li J.H., Wang E.T., Chen W.F., Chen W.X. Genetic diversity and potential for promotion of plant growth detected in nodule endophytic bacteria of soybean grown in Heilongjiang province of China. Soil Biol. Biochem. 2008;40:238–246. doi: 10.1016/j.soilbio.2007.08.014. - DOI
-
- Wang Y., Wang L., Zou Y., Chen L., Cai Z., Zhang S., Zhao F., Tian Y., Jiang Q., Ferguson B.J., et al. Soybean miR172c Targets the Repressive AP2 Transcription Factor NNC1 to Activate ENOD40 Expression and Regulate Nodule Initiation. Plant Cell. 2014;26:4782–4801. doi: 10.1105/tpc.114.131607. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

