In Silico Analysis of Mechanisms of Maribavir-Induced Inhibition and Drug Resistance Mutations in pUL97 Kinase Structural Prediction with AlphaFold2
- PMID: 40733558
- PMCID: PMC12301049
- DOI: 10.3390/v17070941
In Silico Analysis of Mechanisms of Maribavir-Induced Inhibition and Drug Resistance Mutations in pUL97 Kinase Structural Prediction with AlphaFold2
Abstract
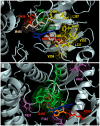
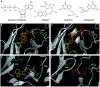
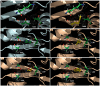
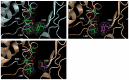
Infections with cytomegalovirus (CMV) can result in increased morbidity and mortality in immunocompromised patients. The pUL97 kinase is a critical enzyme in the regulation of CMV replication. Although it does not phosphorylate deoxynucleosides, this enzyme is involved in the first phosphorylation step of ganciclovir (GCV), a viral DNA polymerase inhibitor. In contrast, maribavir (MBV) is a specific inhibitor of pUL97 kinase activity. In this paper, we analyzed the already-reported amino acid changes, conferring resistance to MBV and cross-resistance to GCV, in the pUL97 protein structure, predicted with AlphaFold2. Docking experiments suggest that MBV is a dual-site inhibitor, targeting ATP binding and substrate phosphorylation. Substitutions that confer resistance to MBV only may directly or indirectly alter the shape of the cavity in the vicinity of the invariant K355 in the putative ATP binding site, without affecting the viral growth. The most frequently encountered T409M substitution may correspond to a gatekeeper mutation. Substitutions that induce cross-resistance to MBV and GCV may directly or indirectly affect the environment of D456 and N461 residues in the catalytic loop, with reduced viral replicative capacity. These results have implications for the clinical use of MBV as well as for the design of novel pUL97 kinase inhibitors.
Keywords: AlphaFold2; cyclopropavir; docking; drug resistance; ganciclovir; gatekeeper residue; human cytomegalovirus; maribavir; pUL97 kinase; predicted protein structure.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Dual Resistance to Maribavir and Ganciclovir in Transplant Recipients.Viruses. 2025 Mar 14;17(3):421. doi: 10.3390/v17030421. Viruses. 2025. PMID: 40143348 Free PMC article.
-
Generation of a panel of mutants that are resistant to standard of care therapies in a clinically relevant strain of human cytomegalovirus for drug resistance profiling.Antiviral Res. 2025 Sep;241:106237. doi: 10.1016/j.antiviral.2025.106237. Epub 2025 Jul 11. Antiviral Res. 2025. PMID: 40653174
-
Cytomegalovirus UL97 kinase mutations that confer maribavir resistance.J Infect Dis. 2007 Jul 1;196(1):91-4. doi: 10.1086/518514. Epub 2007 May 17. J Infect Dis. 2007. PMID: 17538888
-
Cytomegalovirus UL97 mutations in the era of ganciclovir and maribavir.Rev Med Virol. 2008 Jul-Aug;18(4):233-46. doi: 10.1002/rmv.574. Rev Med Virol. 2008. PMID: 18383425 Review.
-
Clinical development of letermovir and maribavir: Overview of human cytomegalovirus drug resistance.Antiviral Res. 2019 Mar;163:91-105. doi: 10.1016/j.antiviral.2019.01.011. Epub 2019 Jan 25. Antiviral Res. 2019. PMID: 30690043 Review.
References
-
- Boivin G., Limaye A.P. Goldman-Cecil Medicine. Elsevier; Amsterdam, The Netherlands: 2023. Cytomegalovirus.
-
- Kern E.R., Kushner N.L., Hartline C.B., Williams-Aziz S.L., Harden E.A., Zhou S., Zemlicka J., Prichard M.N. In vitro activity and mechanism of action of methylenecyclopropane analogs of nucleosides against herpesvirus replication. Antimicrob. Agents Chemother. 2005;49:1039–1045. doi: 10.1128/AAC.49.3.1039-1045.2005. - DOI - PMC - PubMed
-
- James S.H., Hartline C.B., Harden E.A., Driebe E.M., Schupp J.M., Engelthaler D.M., Keim P.S., Bowlin T.L., Kern E.R., Prichard M.N. Cyclopropavir inhibits the normal function of the human cytomegalovirus UL97 kinase. Antimicrob. Agents Chemother. 2011;55:4682–4691. doi: 10.1128/AAC.00571-11. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

