Characterization of Gene Expression Suppression by Bovine Coronavirus Non-Structural Protein 1
- PMID: 40733595
- PMCID: PMC12299145
- DOI: 10.3390/v17070978
Characterization of Gene Expression Suppression by Bovine Coronavirus Non-Structural Protein 1
Abstract
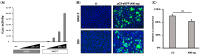
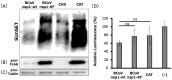
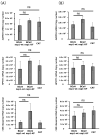
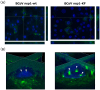
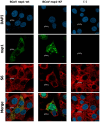
Coronavirus non-structural protein 1 (nsp1) is a pathogenic determinant of Betacoronaviruses. Previous studies demonstrated that the nsp1 of various coronaviruses induces host shutoff through a variety of mechanisms; however, there is little information on the function of bovine coronavirus (BCoV) nsp1. We aimed to characterize the host gene expression suppression function of BCoV nsp1. We first confirmed that the expression of BCoV nsp1 in MAC-T cells, a bovine mammary epithelial cell line, suppressed host and reporter gene expression. Subsequently, lysine and phenylalanine at amino acid positions 232 and 233, respectively, were identified as key residues required for this suppressive effect. Expression levels of housekeeping genes are comparable in cells expressing wild-type BCoV nsp1 and a mutant with alanine substitutions at positions 232 and 233 (BCoV nsp1-KF). Wild-type BCoV nsp1 localized to both the cytoplasm and nucleus; however, BCoV nsp1-KF exhibited prominent nuclear accumulation with dot-like structures. Using confocal microscopy and co-sedimentation analysis, we identified an association between wild-type BCoV nsp1, but not BCoV nsp1-KF, and ribosomes, suggesting that ribosome binding is required for BCoV nsp1-mediated suppression of host gene expression. This is the first study of the characterization of host gene expression suppression by BCoV nsp1.
Keywords: bovine coronavirus; non-structural protein 1; ribosome; shutoff function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The segmented flavivirus Alongshan virus reduces mitochondrial mass by degrading STAT2 to suppress the innate immune response.J Virol. 2025 Jan 31;99(1):e0130124. doi: 10.1128/jvi.01301-24. Epub 2024 Dec 10. J Virol. 2025. PMID: 39655955 Free PMC article.
-
First linear B-cell epitope identified on the nucleocapsid protein of bovine coronavirus.Virology. 2025 Sep;610:110581. doi: 10.1016/j.virol.2025.110581. Epub 2025 May 25. Virology. 2025. PMID: 40483734
-
Detection and characterization of bovine coronavirus and rotavirus in calves in Ethiopia.BMC Vet Res. 2025 Feb 28;21(1):122. doi: 10.1186/s12917-025-04563-9. BMC Vet Res. 2025. PMID: 40022093 Free PMC article.
-
Interventions for fertility preservation in women with cancer undergoing chemotherapy.Cochrane Database Syst Rev. 2025 Jun 19;6(6):CD012891. doi: 10.1002/14651858.CD012891.pub2. Cochrane Database Syst Rev. 2025. PMID: 40536056 Review.
-
Omega-3 fatty acids for depression in adults.Cochrane Database Syst Rev. 2015 Nov 5;2015(11):CD004692. doi: 10.1002/14651858.CD004692.pub4. Cochrane Database Syst Rev. 2015. Update in: Cochrane Database Syst Rev. 2021 Nov 24;11:CD004692. doi: 10.1002/14651858.CD004692.pub5. PMID: 26537796 Free PMC article. Updated.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

