SHOT GLASS, an R2R3-MYB transcription factor, promotes gemma cup and gametangiophore development in Marchantia polymorpha
- PMID: 40734229
- PMCID: PMC12371152
- DOI: 10.1111/nph.70337
SHOT GLASS, an R2R3-MYB transcription factor, promotes gemma cup and gametangiophore development in Marchantia polymorpha
Abstract
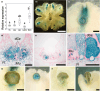
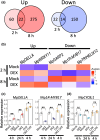
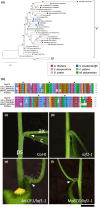
Many plants reproduce asexually by generating clonal progeny from vegetative tissues, a process known as vegetative reproduction. This reproduction mode contrasts with sexual reproduction, which enhances genetic diversity. The bryophyte Marchantia polymorpha L. adjusts its reproductive strategy in response to seasonal environmental cues, transitioning between vegetative and sexual reproduction. In this study, we identified a gene encoding the R2R3-MYB transcription factor SHOT GLASS (MpSTG) as a critical regulator of gemma cup development. MpSTG was predominantly expressed in the gemma cup, apical notch, and sexual reproductive organs (gametangiophores). MpSTG mutation resulted in the formation of abnormal shot-glass-shaped structures lacking gemmae, which replaced functional gemma cups. Additionally, MpSTG-disrupted plants failed to develop sexual reproductive organs, even under inductive conditions. In Arabidopsis thaliana, the MpSTG ortholog LATERAL ORGAN FUSION1 (AtLOF1) plays a pivotal role in lateral bud formation. We demonstrated that MpSTG can partially compensate for AtLOF1's function in lateral bud formation in A. thaliana. Our findings suggest that MpSTG is a key regulator of vegetative and sexual reproduction in M. polymorpha, and illustrate that evolutionarily conserved developmental mechanisms may function in both the gametophyte generation of bryophytes and the sporophyte generation of angiosperms.
Keywords: bryophyte; gametangiophore; gemma cup; organogenesis; sexual reproduction; transcription factor; vegetative reproduction.
© 2025 The Author(s). New Phytologist © 2025 New Phytologist Foundation.
Conflict of interest statement
None declared.
Figures





Similar articles
-
Evolutionary-conserved RLF, a cytochrome b5-like heme-binding protein, regulates organ development in Marchantia polymorpha.New Phytol. 2025 Jul;247(2):929-950. doi: 10.1111/nph.70181. Epub 2025 May 25. New Phytol. 2025. PMID: 40413697 Free PMC article.
-
MpmiR319 promotes gemma/gemma cup formation in the liverwort Marchantia polymorpha.J Exp Bot. 2025 Aug 21;76(12):3378-3389. doi: 10.1093/jxb/eraf148. J Exp Bot. 2025. PMID: 40200901 Free PMC article.
-
GEMMA CUP-ASSOCIATED MYB1, an Ortholog of Axillary Meristem Regulators, Is Essential in Vegetative Reproduction in Marchantia polymorpha.Curr Biol. 2019 Dec 2;29(23):3987-3995.e5. doi: 10.1016/j.cub.2019.10.004. Epub 2019 Nov 7. Curr Biol. 2019. PMID: 31708390
-
Gemma cup and gemma development in Marchantia polymorpha.New Phytol. 2020 Oct;228(2):459-465. doi: 10.1111/nph.16655. Epub 2020 Jun 19. New Phytol. 2020. PMID: 32390245 Review.
-
Building new insights in plant gametogenesis from an evolutionary perspective.Nat Plants. 2019 Jul;5(7):663-669. doi: 10.1038/s41477-019-0466-0. Epub 2019 Jul 8. Nat Plants. 2019. PMID: 31285561 Review.
References
-
- Aki SS, Mikami T, Naramoto S, Nishihama R, Ishizaki K, Kojima M, Takebayashi Y, Sakakibara H, Kyozuka J, Kohchi T et al. 2019. Cytokinin signaling is essential for organ formation in Marchantia polymorpha . Plant and Cell Physiology 60: 1842–1854. - PubMed
-
- Barnes CR, Land WJG. 1908. Bryological papers. II. The origin of the cupule of Marchantia . Botanical Gazette 46: 401–409.
-
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F et al. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171: 287–304. - PubMed
MeSH terms
Substances
Grants and funding
- Yamada Science Foundation
- Ohsumi Frontier Science Foundation
- Kyoto University Foundation
- Suntory Foundation for Life Sciences
- KAKENHI 15H01233/Ministry of Education, Culture, Sports, Science and Technology
- KAKENHI 17H06472/Ministry of Education, Culture, Sports, Science and Technology
- KAKENHI 19H05670/Ministry of Education, Culture, Sports, Science and Technology
- KAKENHI 19H05673/Ministry of Education, Culture, Sports, Science and Technology
- KAKENHI 20H05780/Ministry of Education, Culture, Sports, Science and Technology
- KAKENHI 21K15125/Ministry of Education, Culture, Sports, Science and Technology
- KAKENHI 25119711/Ministry of Education, Culture, Sports, Science and Technology
- Asahi Glass Foundation
- KAKENHI 15H04391/Japan Society for the Promotion of Science
- KAKENHI 19H03247/Japan Society for the Promotion of Science
- KAKENHI 21J40092/Japan Society for the Promotion of Science
- KAKENHI25K09689/Japan Society for the Promotion of Science
- JPMJGX23B0/Japan Science and Technology Agency
- J-PEAKS/Japan Society for the Promotion of Science
LinkOut - more resources
Full Text Sources

