Robust consensus nuclear and cell segmentation
- PMID: 40734708
- PMCID: PMC12303819
- DOI: 10.3389/fgene.2025.1547788
Robust consensus nuclear and cell segmentation
Abstract
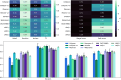
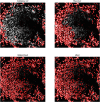
Cell segmentation is a crucial step in numerous biomedical imaging endeavors-so much so that the community is flooded with publicly available, state-of-the-art segmentation techniques ready for out-of-the-box use. Assessing the strengths and limitations of each method on a tissue sample set and then selecting the optimal method for each research objective and input image are time-consuming and exacting tasks that often monopolize the resources of biologists, biochemists, immunologists, and pathologists, despite not being the primary goal of their research projects. In this work, we present a segmentation software wrapper, coined CellSampler, which runs a selection of established segmentation methods and then combines their individual segmentation masks into a single optimized mask. This so-called "uber mask" selects the best of the established masks across local neighborhoods within the image, where both the neighborhood size and the statistical measure used to define what qualifies as "best" are user-defined.
Keywords: bioinformatics; computer vision; imaging mass cytometry; multiplexed imaging; single-cell segmentation.
Copyright © 2025 Irfan, González-Solares, Whitmarsh, Molaeinezhad, Al Sa’d, Mulvey, Ribes, Fatemi, Bressan and Walton.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









References
-
- Boiangiu C.-A., Ioanitescu R. (2013). Journal of information systems and operations management, 1–11Copyright. Copyright Romanian American University Scientific Research Department Winter.
LinkOut - more resources
Full Text Sources

