Evaluating the utility of deep genome skimming for phylogenomic analyses: A case study in the species-rich genus Rhododendron
- PMID: 40734829
- PMCID: PMC12302500
- DOI: 10.1016/j.pld.2025.04.006
Evaluating the utility of deep genome skimming for phylogenomic analyses: A case study in the species-rich genus Rhododendron
Abstract
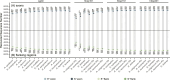
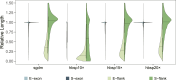
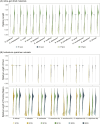
Deep genome skimming (DGS) has emerged as a promising approach to recover orthologous nuclear genes for large-scale phylogenomic analyses. However, its reliability with low DNA quality and quantity typical of archival specimens, such as herbarium material, remains largely unexplored. We used Rhododendron as a case study to evaluate best practices for DGS in phylogenetic analyses at both deep and shallow scales. We first investigated locus recovery variation with sequencing depth, before evaluating the phylogenetic utility of different sets of loci, including Angiosperms353, target nuclear exons, and extended exon-flanking regions. We found DGS effectively recovered nuclear genes from herbarium specimens, with ∼15× coverage performing similarly to deeper sequencing. The recovery of target exon and flanking regions was improved by using supercontigs as a reference, offering a potential solution to limited sequencing depth. The high-integrity nuclear sequences recovered robust phylogenetic relationships within Rhododendron. Notably, exon-flanking regions showed significant potential for resolving relationships at shallow scales. Genes recovered with taxon-specific references had less missing data than those recovered by Angiosperms353 and generated higher-resolution phylogenetic trees. This study demonstrates the utility of DGS data for obtaining numerous nuclear genes from herbarium specimens for phylogenetic studies, and makes recommendations for best practices regarding sequencing coverage, locus selection, and bioinformatic approaches.
Keywords: Deep genome skimming; Degraded DNA; Herbarium specimens; Non-targeted exon-flanking regions; Phylogenomics; Target enrichment.
© 2025 Kunming Institute of Botany, Chinese Academy of Sciences. Publishing services by Elsevier B.V. on behalf of KeAi Communications Co., Ltdé.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Aguirre-Santoro J., Zuluaga A., Stonesmyth E., et al. Phylogenomics of Puya (Bromeliaceae): evolution in the Andean slopes and sky island ecosystems. J. Syst. Evol. 2024;62:257–274.
-
- Bakker F.T., Lei D., Yu J., et al. Herbarium genomics: plastome sequence assembly from a range of herbarium specimens using an iterative organelle genome assembly pipeline. Biol. J. Linn. Soc. 2016;117:33–43.
-
- Burbano H.A., Gutaker R.M. Ancient DNA genomics and the renaissance of herbaria. Science. 2023;382:59–63. - PubMed
LinkOut - more resources
Full Text Sources

